+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4673 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human U5.U4/U6 tri-snRNP, multi-body refinement, core map | |||||||||
 Map data Map data | Human U5.U4/U6 tri-snRNP, multi-body refinement, core map | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
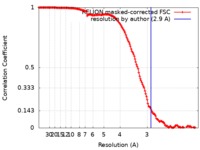
| Method | single particle reconstruction / cryo EM / Resolution: 2.9 Å | |||||||||
 Authors Authors | Charenton C / Wilkinson ME / Nagai K | |||||||||
 Citation Citation |  Journal: Science / Year: 2019 Journal: Science / Year: 2019Title: Mechanism of 5' splice site transfer for human spliceosome activation. Authors: Clément Charenton / Max E Wilkinson / Kiyoshi Nagai /  Abstract: The prespliceosome, comprising U1 and U2 small nuclear ribonucleoproteins (snRNPs) bound to the precursor messenger RNA 5' splice site (5'SS) and branch point sequence, associates with the U4/U6.U5 ...The prespliceosome, comprising U1 and U2 small nuclear ribonucleoproteins (snRNPs) bound to the precursor messenger RNA 5' splice site (5'SS) and branch point sequence, associates with the U4/U6.U5 tri-snRNP to form the fully assembled precatalytic pre-B spliceosome. Here, we report cryo-electron microscopy structures of the human pre-B complex captured before U1 snRNP dissociation at 3.3-angstrom core resolution and the human tri-snRNP at 2.9-angstrom resolution. U1 snRNP inserts the 5'SS-U1 snRNA helix between the two RecA domains of the Prp28 DEAD-box helicase. Adenosine 5'-triphosphate-dependent closure of the Prp28 RecA domains releases the 5'SS to pair with the nearby U6 ACAGAGA-box sequence presented as a mobile loop. The structures suggest that formation of the 5'SS-ACAGAGA helix triggers remodeling of an intricate protein-RNA network to induce Brr2 helicase relocation to its loading sequence in U4 snRNA, enabling Brr2 to unwind the U4/U6 snRNA duplex to allow U6 snRNA to form the catalytic center of the spliceosome. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4673.map.gz emd_4673.map.gz | 260.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4673-v30.xml emd-4673-v30.xml emd-4673.xml emd-4673.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4673_fsc.xml emd_4673_fsc.xml | 14.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_4673.png emd_4673.png | 220.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4673 http://ftp.pdbj.org/pub/emdb/structures/EMD-4673 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4673 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4673 | HTTPS FTP |
-Related structure data
| Related structure data |  4658C  4665C  4672C  4674C  4675C  4676C  4686C  4687C  4688C  4689C  4690C  6qw6C  6qx9C C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10307 (Title: Human pre-B spliceosome and U4/U6.U5 tri-snRNP / Data size: 2.3 TB EMPIAR-10307 (Title: Human pre-B spliceosome and U4/U6.U5 tri-snRNP / Data size: 2.3 TBData #1: Dataset 1 of human pre-B spliceosome; motion-corrected micrographs [micrographs - single frame] Data #2: Dataset 2 of human pre-B spliceosome; motion-corrected micrographs [micrographs - single frame] Data #3: Dataset 3 of human pre-B spliceosome; motion-corrected micrographs [micrographs - single frame] Data #4: Dataset 4 of human pre-B spliceosome; motion-corrected micrographs [micrographs - single frame] Data #5: Dataset 5 of human pre-B spliceosome; motion-corrected micrographs [micrographs - single frame] Data #6: Selected U4/U6.U5 tri-snRNP particles after Bayesian polishing [picked particles - single frame - processed] Data #7: Crude shifted preB particles [picked particles - single frame - unprocessed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4673.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4673.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Human U5.U4/U6 tri-snRNP, multi-body refinement, core map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.022 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Human U4/U6.U5 tri-snRNP
| Entire | Name: Human U4/U6.U5 tri-snRNP |
|---|---|
| Components |
|
-Supramolecule #1: Human U4/U6.U5 tri-snRNP
| Supramolecule | Name: Human U4/U6.U5 tri-snRNP / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#32 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.7 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.3 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.9 Component:
| ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK III / Details: Wait 30s, blot for 2s to 3s.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 1-40 / Average exposure time: 6.0 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)