+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3409 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM of nanoscale DNA assemblies | |||||||||
 Map data Map data | Reconstruction of DNA icosahedron origami | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Scaffolded DNA origami | |||||||||
| Biological species |  Enterobacteria phage M13 (virus) / synthetic construct (others) Enterobacteria phage M13 (virus) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 20.0 Å | |||||||||
 Authors Authors | Zhang K / Chiu W / Veneziano R / Ratanalert S / Zhang F / Yan H / Bathe M | |||||||||
 Citation Citation |  Journal: Science / Year: 2016 Journal: Science / Year: 2016Title: Designer nanoscale DNA assemblies programmed from the top down. Authors: Rémi Veneziano / Sakul Ratanalert / Kaiming Zhang / Fei Zhang / Hao Yan / Wah Chiu / Mark Bathe /  Abstract: Scaffolded DNA origami is a versatile means of synthesizing complex molecular architectures. However, the approach is limited by the need to forward-design specific Watson-Crick base pairing manually ...Scaffolded DNA origami is a versatile means of synthesizing complex molecular architectures. However, the approach is limited by the need to forward-design specific Watson-Crick base pairing manually for any given target structure. Here, we report a general, top-down strategy to design nearly arbitrary DNA architectures autonomously based only on target shape. Objects are represented as closed surfaces rendered as polyhedral networks of parallel DNA duplexes, which enables complete DNA scaffold routing with a spanning tree algorithm. The asymmetric polymerase chain reaction is applied to produce stable, monodisperse assemblies with custom scaffold length and sequence that are verified structurally in three dimensions to be high fidelity by single-particle cryo-electron microscopy. Their long-term stability in serum and low-salt buffer confirms their utility for biological as well as nonbiological applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3409.map.gz emd_3409.map.gz | 48.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3409-v30.xml emd-3409-v30.xml emd-3409.xml emd-3409.xml | 16.3 KB 16.3 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-3409.png EMD-3409.png | 203.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3409 http://ftp.pdbj.org/pub/emdb/structures/EMD-3409 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3409 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3409 | HTTPS FTP |
-Validation report
| Summary document |  emd_3409_validation.pdf.gz emd_3409_validation.pdf.gz | 227.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3409_full_validation.pdf.gz emd_3409_full_validation.pdf.gz | 227.1 KB | Display | |
| Data in XML |  emd_3409_validation.xml.gz emd_3409_validation.xml.gz | 6.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3409 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3409 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3409 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3409 | HTTPS FTP |
-Related structure data
| Related structure data | 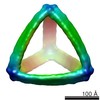 3408C  3410C  3411C  3412C  3413C C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10068 (Title: Designer nanoscale DNA assemblies programmed from the top down EMPIAR-10068 (Title: Designer nanoscale DNA assemblies programmed from the top downData size: 13.0 / Data #1: DNA icosahedron [micrographs - single frame]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3409.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3409.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of DNA icosahedron origami | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.51 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : DNA icosahedron, 52-bp edge length
| Entire | Name: DNA icosahedron, 52-bp edge length |
|---|---|
| Components |
|
-Supramolecule #1000: DNA icosahedron, 52-bp edge length
| Supramolecule | Name: DNA icosahedron, 52-bp edge length / type: sample / ID: 1000 Details: Monodisperse and pure sample were prepared by overnight folding reaction and purified using centrifugal filter (MWCO 100KDa) Oligomeric state: monomer / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 2.07 MDa / Theoretical: 2.07 MDa / Method: Calculation |
-Macromolecule #1: M13mp18 phage genome segment
| Macromolecule | Name: M13mp18 phage genome segment / type: dna / ID: 1 / Name.synonym: Scaffold strand / Details: amplified using assymetric PCR / Classification: DNA / Structure: SINGLE STRANDED / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage M13 (virus) / Strain: M13mp18 / synonym: M13 phage Enterobacteria phage M13 (virus) / Strain: M13mp18 / synonym: M13 phage |
| Molecular weight | Experimental: 1.01 MDa / Theoretical: 1.01 MDa |
| Sequence | String: TCTTTGCCTT GCCTGTATGA TTTATTGGAT GTTAATGCTA CTACTATTAG TAGAATTGAT GCCACCTTTT CAGCTCGCGC CCCAAATGAA AATATAGCTA AACAGGTTAT TGACCATTTG CGAAATGTAT CTAATGGTCA AACTAAATCT ACTCGTTCGC AGAATTGGGA ...String: TCTTTGCCTT GCCTGTATGA TTTATTGGAT GTTAATGCTA CTACTATTAG TAGAATTGAT GCCACCTTTT CAGCTCGCGC CCCAAATGAA AATATAGCTA AACAGGTTAT TGACCATTTG CGAAATGTAT CTAATGGTCA AACTAAATCT ACTCGTTCGC AGAATTGGGA ATCAACTGTT ACATGGAATG AAACTTCCAG ACACCGTACT TTAGTTGCAT ATTTAAAACA TGTTGAGCTA CAGCACCAGA TTCAGCAATT AAGCTCTAAG CCATCCGCAA AAATGACCTC TTATCAAAAG GAGCAATTAA AGGTACTCTC TAATCCTGAC CTGTTGGAGT TTGCTTCCGG TCTGGTTCGC TTTGAAGCTC GAATTAAAAC GCGATATTTG AAGTCTTTCG GGCTTCCTCT TAATCTTTTT GATGCAATCC GCTTTGCTTC TGACTATAAT AGTCAGGGTA AAGACCTGAT TTTTGATTTA TGGTCATTCT CGTTTTCTGA ACTGTTTAAA GCATTTGAGG GGGATTCAAT GAATATTTAT GACGATTCCG CAGTATTGGA CGCTATCCAG TCTAAACATT TTACTATTAC CCCCTCTGGC AAAACTTCTT TTGCAAAAGC CTCTCGCTAT TTTGGTTTTT ATCGTCGTCT GGTAAACGAG GGTTATGATA GTGTTGCTCT TACTATGCCT CGTAATTCCT TTTGGCGTTA TGTATCTGCA TTAGTTGAAT GTGGTATTCC TAAATCTCAA CTGATGAATC TTTCTACCTG TAATAATGTT GTTCCGTTAG TTCGTTTTAT TAACGTAGAT TTTTCTTCCC AACGTCCTGA CTGGTATAAT GAGCCAGTTC TTAAAATCGC ATAAGGTAAT TCACAATGAT TAAAGTTGAA ATTAAACCAT CTCAAGCCCA ATTTACTACT CGTTCTGGTG TTCTCGTCAG GGCAAGCCTT ATTCACTGAA TGAGCAGCTT TGTTACGTTG ATTTGGGTAA TGAATATCCG GTTCTTGTCA AGATTACTCT TGATGAAGGT CAGCCAGCCT ATGCGCCTGG TCTGTACACC GTTCATCTGT CCTCTTTCAA AGTTGGTCAG TTCGGTTCCC TTATGATTGA CCGTCTGCGC CTCGTTCCGG CTAAGTAACA TGGAGCAGGT CGCGGATTTC GACACAATTT ATCAGGCGAT GATACAAATC TCCGTTGTAC TTTGTTTCGC GCTTGGTATA ATCGCTGGGG GTCAAAGATG AGTGTTTTAG TGTATTCTTT CGCCTCTTTC GTTTTAGGTT GGTGCCTTCG TAGTGGCATT ACGTATTTTA CCCGTTTAAT GGAAACTTCC TCATGAAAAA GTCTTTAGTC CTCAAAGCCT CTGTAGCCGT TGCTACCCTC GTTCCGATGC TGTCTTTCGC TGCTGAGGGT GACGATCCCG CAAAAGCGGC CTTTAACTCC CTGCAAGCCT CAGCGACCGA ATATATCGGT TATGCGTGGG CGATGGTTGT TGTCATTGTC GGCGCAACTA TCGGTATCAA GCTGTTTAAG AAATTCACCT CGAAAGCAAG CTGATAAACC GATACAATTA AAGGCTCCTT TTGGAGCCTT TTTTTTTGGA GATTTTCAAC GTGAAAAAAT TATTATTCGC AATTCCTTTA GTTGTTCCTT TCTATTCTCA CTCCGCTGAA ACTGTTGAAA GTTGTTTAGC AAAACCCCAT ACAGAAAATT CATTTACTAA CGTCTGGAAA GACGACAAAA CTTTAGATCG TTACGCTAAC TATGAGGGTT GTCTGTGGAA TGCTACAGGC GTTGTAGTTT GTACTGGTGA CGAAACTCAG TGTTACGGTA CATGGGTTCC TATTGGGCTT GCTATCCCTG AAAATGAGGG TGGTGGCTCT GAGGGTGGCG GTTCTGAGGG TGGCGGTTCT GAGGGTGGCG GTACTAAACC TCCTGAGTAC GGTGATACAC CTATTCCGGG CTATACTTAT ATCAACCCTC TCGACGGCAC TTATCCGCCT GGTACTGAGC AAAACCCCGC TAATCCTAAT CCTTCTCTTG AGGAGTCTCA GCCTCTTAAT ACTTTCATGT TTCAGAATAA TAGGTTCCGA AATAGGCAGG GGGCATTAAC TGTTTATACG GGCACTGTTA CTCAAGGCAC TGACCCCGTT AAAACTTATT ACCAGTACAC TCCTGTATCA TCAAAAGCCA TGTATGACGC TTACTGGAAC GGTAAATTCA GAGACTGCGC TTTCCATTCT GGCTTTAATG AAGATCCATT CGTTTGTGAA TATCAAGGCC AATCGTCTGA CCTGCCTCAA CCTCCTGTCA ATGCTGGCGG CGGCTCTGGT GGTGGTTCTG GTGGCGGCTC TGAGGGTGGT GGCTCTGAGG GTGGCGGTTC TGAGGGTGGC GGCTCTGAGG GAGGCGGTTC CGGTGGTGGC TCTGGTTCCG GTGATTTTGA TTATGAAAAG ATGGCAAACG CTAATAAGGG GGCTATGACC GAAAATGCCG ATGAAAACGC GCTACAGTCT GACGCTAAAG GCAAACTTGA TTCTGTCGCT ACTGATTACG GTGCTGCTAT CGATGGTTTC ATTGGTGACG TTTCCGGCCT TGCTAATGGT AATGGTGCTA CTGGTGATTT TGCTGGCTCT AATTCCCAAA TGGCTCAAGT CGGTGACGGT GATAATTCAC CTTTAATGAA TAATTTCCGT CAATATTTAC CTTCCCTCCC TCAATCGGTT GAATGTCGCC CTTTTGTCTT TAGCGCTGGT AAACCATATG AATTTTCTAT TGATTGTGAC AAAATAAACT TATTCCGTGG TGTCTTTGCG TTTCTTTTAT ATGTTGCCAC CTTTATGTAT GTATTTTCTA CGTTTGCTAA CATACTGCGT AATAAGGAGT CTTAATCATG CCAGTTCTTT TGGGTATTCC GTTATTATTG CGTTTCCTCG GTTTCCTTCT GGTAACTTTG TTCGGCTATC TGCTTACTTT TCTTAAAAAG GGCTTCGGTA AGATAGCTAT TGCTATTTCA TTGTTTCTTG CTCTTATTAT TGGGCTTAAC TCAATTCTTG TGGGTTATCT CTCTGATATT AGCGCTCAAT TACCCTCTGA CTTTGTTCAG GGTGTTCAGT TAATTCTCCC GTCTAATGCG CTTCCCTGTT TTTATGTTAT TCTCTCTGTA AAGGCTGCTA TTTTCATTTT TGACGTTAAA CAAAAAATCG TTTCTTATTT GGATTGGGAT AAATAATATG GCTGTTTATT TTGTAACTGG CAAATTAGGC TCTGGAAAGA CGCTCGTTAG C |
-Macromolecule #2: Synthetic DNA oligonucleotides
| Macromolecule | Name: Synthetic DNA oligonucleotides / type: dna / ID: 2 / Name.synonym: Staple strand Details: CAAGAACCGGATTTTTTATTCATTACGGATCTTCATTTTTTTAAAGCCAGAA CTTGATATTCGTAACAAAGCTGCTCATTCAGAGACGATTGGC ACAAACGAATCCAAATCAAC GATTTTAATAATCATTGTGAATTACAAGAGT AATCTTGACTGGCTGACCTTCATCCTTATGC ...Details: CAAGAACCGGATTTTTTATTCATTACGGATCTTCATTTTTTTAAAGCCAGAA CTTGATATTCGTAACAAAGCTGCTCATTCAGAGACGATTGGC ACAAACGAATCCAAATCAAC GATTTTAATAATCATTGTGAATTACAAGAGT AATCTTGACTGGCTGACCTTCATCCTTATGC TGAATAAGGCTTTTTTTGCCCTGACGACGATAAAAACTTTTTCAAAATAGCG AATTACGAGGCTTTTTATAGTAAGAGGAGATGGTTTATTTTTATTTCAACTT ACGAGTAGTAATAACCCTCGTTTACCAGACGAGAACACCAGA AATTGGGCTTCAACACTATC GAATCGTCGACTGGATAGCGTCCACATAACG CCAAAAGGCAACTAATGCAGATAATACTGCG ATATCGCGTTTTTTTTTAATTCGAGCGGGGTAATAGTTTTTTAAAATGTTTA CAAAAGAAGTACCAGACCGGAAGCAAACTCCAGAGGCTTTTG TTTGCCAGAGTTCAAAGCGA GCTCAACATTAATTGCTGAATCTGCCCGAAA GACTTCAAAAGATTAAGAGGAAGGTGCTGTA CGAGCTGAAAATTTTTGGTGGCATCATTTTTGCGGATTTTTTGGCTTAGAGC CTCCTTTTGATAGTAGTAGCATTAGAATAACACCTTTAATTG TAAGAGGTCAATTCTACTAA AGAATTGACTAATATCAGAGAGATTTTTCAT TTGGGGCGACCTGTTTAGCTATAAACCCACA AACAGGTCAGGTTTTTATTAGAGAGTATAAAAACAGGTTTTTGAAGCGCATTGACAGGAGGTTTTTTTGAGGCAGGTC TTAACTGAACACCAGAGCCGCCGCCAGCATTAGACGGGAGAA ACCCTGAACACAGAACCACC ACCGGAACCGCTTTTTCTCCCTCAGACACCACCCTCATTTTTGAGCCGCCAC CTCAGAGCGCCGCCACCCTCAGAACTGAATT TACCGTTCTGGAAAGCGCAGTCTCCGCCACC CGGGATCGTCATTTTTCCCTCAGCAGTTTTAACGGGGTTTTTTCAGTGCCTTCCCTGCCTATTTTTTTTCGGAACCTA TTAATGCCGAGTAACAGTGCCCGTGGAACCA GAGCCACCATAATCAAAATCACCATAAACAG AGGAGTGTACCATCGGAACGAGGGTAGCAACTTTGATGATAC TGGTAATAAGCGAAAGACAG CAGTAAGCGTCTTTTTATACATGGCTGGCTACAGAGGTTTTTCTTTGAGGACTGTACAGACCATTTTTGGCGCATAGG GTCGAAATCCGTTTTTCGACCTGCTCTCAATCATAAGTTTTTGGAACCGAACAACGGGTAAAATTTTTTACGTAATGC TCATGAGGAATGAAAGAGGACAGATGAACGGTAAAGACTTTT GTTTCCATTATGACCAACTT GCAGACGGCATGTTACTTAGCCGGAAAAATC TACGTTAACAGGACGTTGGGAAGAACGAGGC AAGGAATTAGTGAGAATAGAAAGGCGCCTGA TAAATTGTGGAGATTTGTATCATAACAACTA GAACTGGCTCATTTTTTTATACCAGTTAAAACGAACTTTTTTAACGGAACAAGAGATTTAGGATTTTTATACCACATT TGCTAAACAAATGAATTTTCTGTAAAAGATT CATCAGTTCATTATTACAGGTAGTGGGGTTT CCTGTAGCATTTTTTTCCACAGACAAGTCGTCTTTCCTTTTTAGACGTTAGT AAAGTTTTCCCTCATAGTTAGCGTCAGTTCA GAAAACGACTCAAATGCTTTAAAAACGATCT AGGGCGACTGGTTTACCAGCGCTAGTACAAA CTACAACGTGAGTTTCGTCACCAAAGACAAA ATAAATATTCATTTTTTTGAATCCCCGAATGACCATATTTTTAATCAAAAATAGCAAAGCGGATTTTTTTGCATCAAA ATTTTGTCGCAAAGACACCACGGAACTATTA TAGTCAGACAGGTCTTTACCCTGATAAGTTT AAGACTCCTTATTTTTTTACGCAGTAGTGGCAACATATTTTTTAAAAGAAAC ACATAAAGTGTTAGCAAACGTAGAGTTTCAT TCCATGTAGTACGGTGTCTGGAAAAATACAT CCGAAGCCCTTTTTTTTTTAAGAAAAGAAACCGAGGATTTTTAACGCAATAAAAAATCACCAGTTTTTTAGCACCATT ATTTGGGAATCCCAAAAGAACTGGCATGATTCGACTTGAGCC TAGAGCCAGCTAACGGAATA TGTTTTAAATATTTTTTGCAACTAAAACAGTTGATTCTTTTTCCAATTCTGCACATTTCGCAATTTTTATGGTCAATA ACCAGAAGGTAAGCAGATAGCCGAGTTTGAC CATTAGATGAACGAGTAGATTTAACAAAGTT AAGTCAGAGGGTTTTTTAATTGAGCGGTTAAGCCCAATTTTTTAATAAGAGCATTAGCGTTTGTTTTTCCATCTTTTC AAATAGCAATATTTTCGGTCATAGCCCCCTTAAGAAACAATG AGCTATCTTATTTCATCGGC CGCTTTTGTGAGGCTTGCAGGGAGAACCTAA AACGAAAGCACTACGAAGGCACCTTAAAGGC TGAGACTCCTCTTTTTAAGAGAAGGAATAACCGATATTTTTTATTCGGTCGC ACAGCTTGATATTTTTCCGATAGTTGGCGGATAAGTGTTTTTCCGTCGAGAG GGGGTTTTGCGACAACAACCATCGCCCACGCTTAGGATTAGC TCAGTACCAGCGCCGACAAT AGGCGAAAGAATTTTTTACACTAAAAAAGCGCGAAACTTTTTAAAGTACAAC TTTCTTAATTATCAGCTTGCTTTCCCCAGCG ATTATACCCACTCATCTTTGACCGAGGTGAA GAAACCATCGATTTTTTAGCAGCACCTAGCGTCAGACTTTTTTGTAGCGCGT TTTGCCTTGTAATCAGTAGCGACAAAAGTAT TAAGAGGCTTATTCTGAAACATGGAATCAAG TAGGTGTAGGTTGATATAAGTATAGGAAACG TCACCAATACCATTAGCAAGGCCGCCCGGAA AACTTTCAACATTTTTGTTTCAGCGGGCGAATAATAATTTTTTTTTTTCACGGGAACCCATGTTTTTTACCGTAACAC TCACCGTACTCTTTTTAGGAGGTTTAACCGCCACCCTTTTTTCAGAGCCACCGGAGCCTTTAATTTTTTTGTATCGGT CAAAAAAAAATCAGGGATAGCAAGCCCAATATTGAAAATCTC GGCTCCAAAAACCCTCATTT ACAATCAATAGTTTTTAAAATTCATAATTCAACCGATTTTTTTGAGGGAGGGAAGGTGAATTATTTTTTCACCGTCAC CCCTCAGAGTACCGCCACCCTCAGGGAAATT ATTCATTAAAGGTAAATATTGACAACCGCCA Classification: DNA / Structure: SINGLE STRANDED / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Experimental: 1.06 MDa / Theoretical: 1.06 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.05 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 40 mM Tris-HCl, 20 mM acetic acid, 2 mM EDTA |
| Grid | Details: glow-discharged 200-mesh R1.2/1.3 Quantifoil grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV / Method: Blot for 1.5 second before plunging |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2200FS |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 80,000 times magnification |
| Specialist optics | Energy filter - Name: JEOL |
| Details | 5k * 4k |
| Date | Oct 5, 2015 |
| Image recording | Category: CCD / Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Number real images: 180 / Average electron dose: 63 e/Å2 Details: Every image is the average of 60 frames recorded by the direct electron detecto |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 4.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 25000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Details | The particles were selected manually using EMAN2 |
|---|---|
| CTF correction | Details: each particle |
| Final reconstruction | Applied symmetry - Point group: I (icosahedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 20.0 Å / Resolution method: OTHER / Software - Name: EMAN2 / Number images used: 3650 |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















