+Search query
-Structure paper
| Title | Designer nanoscale DNA assemblies programmed from the top down. |
|---|---|
| Journal, issue, pages | Science, Vol. 352, Issue 6293, Page 1534, Year 2016 |
| Publish date | Jun 24, 2016 |
 Authors Authors | Rémi Veneziano / Sakul Ratanalert / Kaiming Zhang / Fei Zhang / Hao Yan / Wah Chiu / Mark Bathe /  |
| PubMed Abstract | Scaffolded DNA origami is a versatile means of synthesizing complex molecular architectures. However, the approach is limited by the need to forward-design specific Watson-Crick base pairing manually ...Scaffolded DNA origami is a versatile means of synthesizing complex molecular architectures. However, the approach is limited by the need to forward-design specific Watson-Crick base pairing manually for any given target structure. Here, we report a general, top-down strategy to design nearly arbitrary DNA architectures autonomously based only on target shape. Objects are represented as closed surfaces rendered as polyhedral networks of parallel DNA duplexes, which enables complete DNA scaffold routing with a spanning tree algorithm. The asymmetric polymerase chain reaction is applied to produce stable, monodisperse assemblies with custom scaffold length and sequence that are verified structurally in three dimensions to be high fidelity by single-particle cryo-electron microscopy. Their long-term stability in serum and low-salt buffer confirms their utility for biological as well as nonbiological applications. |
 External links External links |  Science / Science /  PubMed:27229143 / PubMed:27229143 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 20.0 - 40.0 Å |
| Structure data | 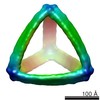 EMDB-3408:  EMDB-3409:  EMDB-3410:  EMDB-3411:  EMDB-3412:  EMDB-3413: |
| Source |
|
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About Yorodumi Papers
About Yorodumi Papers



 Enterobacteria phage M13 (virus)
Enterobacteria phage M13 (virus)