+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30440 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM analysis of the nonribosomal peptide synthetase, FmoA3 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationamino acid activation for nonribosomal peptide biosynthetic process / secondary metabolite biosynthetic process / lipid biosynthetic process / ligase activity / phosphopantetheine binding / antibiotic biosynthetic process / nucleotide binding / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Streptomyces sp. Sp080513GE-23 (bacteria) Streptomyces sp. Sp080513GE-23 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.55 Å | |||||||||
 Authors Authors | Sone K / Harada A / Kawai S / Urano N / Adachi N / Moriya T / Kawasaki M / Katsuyama Y / Senda T / Ohnishi Y | |||||||||
| Funding support |  Japan, 2 items Japan, 2 items
| |||||||||
 Citation Citation |  Journal: Angew Chem Int Ed Engl / Year: 2021 Journal: Angew Chem Int Ed Engl / Year: 2021Title: Structural and Functional Analyses of the Tridomain-Nonribosomal Peptide Synthetase FmoA3 for 4-Methyloxazoline Ring Formation. Authors: Yohei Katsuyama / Kaoru Sone / Ayaka Harada / Seiji Kawai / Naoki Urano / Naruhiko Adachi / Toshio Moriya / Masato Kawasaki / Kazuo Shin-Ya / Toshiya Senda / Yasuo Ohnishi /  Abstract: Nonribosomal peptide synthetases (NRPSs) are attractive targets for bioengineering to generate useful peptides. FmoA3 is a single modular NRPS composed of heterocyclization (Cy), adenylation (A), and ...Nonribosomal peptide synthetases (NRPSs) are attractive targets for bioengineering to generate useful peptides. FmoA3 is a single modular NRPS composed of heterocyclization (Cy), adenylation (A), and peptidyl carrier protein (PCP) domains. It uses α-methyl-l-serine to synthesize a 4-methyloxazoline ring, probably with another Cy domain in the preceding module FmoA2. Here, we determined the head-to-tail homodimeric structures of FmoA3 by X-ray crystallography (apo-form, with adenylyl-imidodiphosphate and α-methyl-l-seryl-AMP) and cryogenic electron microscopy single particle analysis, and performed site-directed mutagenesis experiments. The data revealed that α-methyl-l-serine can be accommodated in the active site because of the extra space around Ala688. The Cy domains of FmoA2 and FmoA3 catalyze peptide bond formation and heterocyclization, respectively. FmoA3's Cy domain seems to lose its donor PCP binding activity. The collective data support a proposed catalytic cycle of FmoA3. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30440.map.gz emd_30440.map.gz | 391.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30440-v30.xml emd-30440-v30.xml emd-30440.xml emd-30440.xml | 18.7 KB 18.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30440_fsc.xml emd_30440_fsc.xml | 17 KB | Display |  FSC data file FSC data file |
| Images |  emd_30440.png emd_30440.png | 125.5 KB | ||
| Others |  emd_30440_half_map_1.map.gz emd_30440_half_map_1.map.gz emd_30440_half_map_2.map.gz emd_30440_half_map_2.map.gz | 337.2 MB 337.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30440 http://ftp.pdbj.org/pub/emdb/structures/EMD-30440 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30440 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30440 | HTTPS FTP |
-Related structure data
| Related structure data |  6ltaC  6ltbC  6ltcC  6ltdC C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-11059 (Title: Structural and functional analyses of the tridomain-nonribosomal peptide synthetase FmoA3 for 4-methyloxazoline ring formation EMPIAR-11059 (Title: Structural and functional analyses of the tridomain-nonribosomal peptide synthetase FmoA3 for 4-methyloxazoline ring formationData size: 1.4 TB Data #1: Structural and functional analyses of the tridomain-nonribosomal peptide synthetase FmoA3 for 4-methyloxazoline ring formation [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30440.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30440.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.88 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
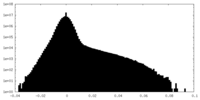
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: #1
| File | emd_30440_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
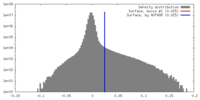
| Density Histograms |
-Half map: #2
| File | emd_30440_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Structure of nonribosomal peptide synthetase, FmoA3
| Entire | Name: Structure of nonribosomal peptide synthetase, FmoA3 |
|---|---|
| Components |
|
-Supramolecule #1: Structure of nonribosomal peptide synthetase, FmoA3
| Supramolecule | Name: Structure of nonribosomal peptide synthetase, FmoA3 / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Streptomyces sp. Sp080513GE-23 (bacteria) Streptomyces sp. Sp080513GE-23 (bacteria) |
| Molecular weight | Theoretical: 120 KDa |
| Recombinant expression | Organism:  |
-Macromolecule #1: FmoA3
| Macromolecule | Name: FmoA3 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Streptomyces sp. Sp080513GE-23 (bacteria) Streptomyces sp. Sp080513GE-23 (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MTISVPGHGT LGAPDGAAAP GGEAAELPPL VPEPGDAGQP FPLTPTQQAL WVGRADAVDL GDIGCYGYFE WERPELDLA RYRRAWERLV AHHPGLRTVV RPDGTQHVLE RPGPVPITVE DLRQDPDAVR RLEESRAERG H RALDPGTW PMFDLRVVLL SGRVRVQLGI ...String: MTISVPGHGT LGAPDGAAAP GGEAAELPPL VPEPGDAGQP FPLTPTQQAL WVGRADAVDL GDIGCYGYFE WERPELDLA RYRRAWERLV AHHPGLRTVV RPDGTQHVLE RPGPVPITVE DLRQDPDAVR RLEESRAERG H RALDPGTW PMFDLRVVLL SGRVRVQLGI DLQLMDASSL FLNLFSDLVT LYDDPDAALA SQKLAFRDFA RW LEEDVRG GARWRADWAY WQERLDGLPP APDLPAARYG AQGPGKFERC MVRCPAEEFA LLRERALAHG LTE TELLVG AFAEVLRGWS SDPAFTLNVP VFQRFDVPGI EDVIGDYTNP ILLEARPEGR TVAERIVALA ARLR ADTRH ASVNGVEVLR ELARRRGLAA AAMPVVVTSL LGLPSAARSI TEFGTEVHSI TQTPQVSLDF QIRPE DGEL RLVWDHRSGA FAPGVVEGAF EAFLDLVGRM LADEPGHGVW EAPFADMRSR RDRAVWNETN DTAEPV PAV LLQERFFAQA RRTPDAEAVV ASGLRLTYDE LARHAYRIGN TLRERGVRPG DLVGVVMEKG WEQYAAV YG ILAAGGAYLP IDAASPRGRV ARLLESAGAG IVLTQSRLRD ELDLPAGTTV LRADTDFETA STAPLTPV Q GPDDPAYVIY TSGSTGEPKG VVVAHRGVAN LVRDVRRRFA VTPADRLLAL SGLHFDASVY DVFGPLACG ATVVVPPPFR RAEPDVWAEL VRDERVTFWN SVPVLLELLV GEAESRDDRP LATLRLAVVS GDWIPLDLPG RARAQAPGL RVVGSGGPTE TICWSLFHPI DAVDPQWTSI PYGKPIANQR YYIVDRDLRP RPTWARGEMA V ASPLGLAL GYLNDPERTA AKFVTLPGTG ERAYLTGDFG RLLPDGGIEI LGREDFQVKV AGQRIELGEI EA LLHRADG VRAAVVTAPR SSADVVRLQA FVVPETGARL SADALREHLS AELPAAMVPA AIRLLPELPL TAN GKVDRL ALARLAAAPE EAPEPEARTD YAPRTDVGLL AELVAACVAE LLGLDEVPTT GNFFRLGGDS LSGT RLASR LQDLLGAPVP IRTVFGNPVL GDLASAIAGD PAAGPQAIRV ARLLHTLEEP DEKPGEKPDA EPAGE PDAG SRT |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 8 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR / Details: The grid was washed by acetone prior to use. | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV / Details: Blotting time was 15 seconds (blot force 25). | |||||||||
| Details | This sample was mono-disperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 1886 / Average exposure time: 50.07 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 120000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller


















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)