[English] 日本語
 Yorodumi
Yorodumi- EMDB-30413: Cryo-EM structure of K+-bound hERG channel in the presence of ast... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-30413 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of K+-bound hERG channel in the presence of astemizole | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | potassium channel / TRANSPORT PROTEIN | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
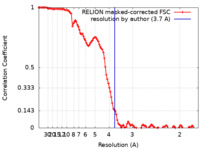
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | ||||||||||||
 Authors Authors | Asai T / Adachi N | ||||||||||||
| Funding support |  Japan, 3 items Japan, 3 items
| ||||||||||||
 Citation Citation |  Journal: Structure / Year: 2021 Journal: Structure / Year: 2021Title: Cryo-EM Structure of K-Bound hERG Channel Complexed with the Blocker Astemizole. Authors: Tatsuki Asai / Naruhiko Adachi / Toshio Moriya / Hideyuki Oki / Takamitsu Maru / Masato Kawasaki / Kano Suzuki / Sisi Chen / Ryohei Ishii / Kazuko Yonemori / Shigeru Igaki / Satoshi Yasuda / ...Authors: Tatsuki Asai / Naruhiko Adachi / Toshio Moriya / Hideyuki Oki / Takamitsu Maru / Masato Kawasaki / Kano Suzuki / Sisi Chen / Ryohei Ishii / Kazuko Yonemori / Shigeru Igaki / Satoshi Yasuda / Satoshi Ogasawara / Toshiya Senda / Takeshi Murata /  Abstract: The hERG channel is a voltage-gated potassium channel involved in cardiac repolarization. Off-target hERG inhibition by drugs has become a critical issue in the pharmaceutical industry. The three- ...The hERG channel is a voltage-gated potassium channel involved in cardiac repolarization. Off-target hERG inhibition by drugs has become a critical issue in the pharmaceutical industry. The three-dimensional structure of the hERG channel was recently reported at 3.8-Å resolution using cryogenic electron microscopy (cryo-EM). However, the drug inhibition mechanism remains unclear because of the scarce structural information regarding the drug- and potassium-bound hERG channels. In this study, we obtained the cryo-EM density map of potassium-bound hERG channel complexed with astemizole, a well-known hERG inhibitor that increases risk of potentially fatal arrhythmia, at 3.5-Å resolution. The structure suggested that astemizole inhibits potassium conduction by binding directly below the selectivity filter. Furthermore, we propose a possible binding model of astemizole to the hERG channel and provide insights into the unusual sensitivity of hERG to several drugs. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_30413.map.gz emd_30413.map.gz | 115 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-30413-v30.xml emd-30413-v30.xml emd-30413.xml emd-30413.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_30413_fsc.xml emd_30413_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_30413.png emd_30413.png | 75 KB | ||
| Filedesc metadata |  emd-30413.cif.gz emd-30413.cif.gz | 6.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-30413 http://ftp.pdbj.org/pub/emdb/structures/EMD-30413 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30413 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-30413 | HTTPS FTP |
-Related structure data
| Related structure data |  7cn1MC  7cn0C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10629 (Title: Cryo-EM structure of K+-bound hERG channel in the presence of astemizole EMPIAR-10629 (Title: Cryo-EM structure of K+-bound hERG channel in the presence of astemizoleData size: 1.8 TB Data #1: Cryo-EM structure of K+-bound hERG channel in the presence of astemizole [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_30413.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_30413.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.884 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : potassium channel
| Entire | Name: potassium channel |
|---|---|
| Components |
|
-Supramolecule #1: potassium channel
| Supramolecule | Name: potassium channel / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: homo-tetramer |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: potassium channel
| Macromolecule | Name: potassium channel / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 91.792086 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MPVRRGHVAP QNTFLDTIIR KFEGQSRKFI IANARVENCA VIYCNDGFCE LCGYSRAEVM QRPCTCDFLH GPRTQRRAAA QIAQALLGA EERKVEIAFY RKDGSCFLCL VDVVPVKNED GAVIMFILNF EVVMEKDMVG SSPTSDREII APKIKERTHN V TEKVTQVL ...String: MPVRRGHVAP QNTFLDTIIR KFEGQSRKFI IANARVENCA VIYCNDGFCE LCGYSRAEVM QRPCTCDFLH GPRTQRRAAA QIAQALLGA EERKVEIAFY RKDGSCFLCL VDVVPVKNED GAVIMFILNF EVVMEKDMVG SSPTSDREII APKIKERTHN V TEKVTQVL SLGADVLPEY KLQAPRIHRW TILHYSPFKA VWDWLILLLV IYTAVFTPYS AAFLLKETEE GPPATECGYA CQ PLAVVDL IVDIMFIVDI LINFRTTYVN ANEEVVSHPG RIAVHYFKGW FLIDMVAAIP FDLLIFGSGS EELIGLLKTA RLL RLVRVA RKLDRYSEYG AAVLFLLMCT FALIAHWLAC IWYAIGNMEQ PHMDSRIGWL HNLGDQIGKP YNSSGLGGPS IKDK YVTAL YFTFSSLTSV GFGNVSPNTN SEKIFSICVM LIGSLMYASI FGNVSAIIQR LYSGTARYHT QMLRVREFIR FHQIP NPLR QRLEEYFQHA WSYTNGIDMN AVLKGFPECL QADICLHLNR SLLQHCKPFR GATKGCLRAL AMKFKTTHAP PGDTLV HAG DLLTALYFIS RGSIEILRGD VVVAILGKND IFGEPLNLYA RPGKSNGDVR ALTYCDLHKI HRDDLLEVLD MYPEFSD HF WSSLEITFNL RDTNMIPGGR QYQELPRCPA PTPSLLNIPL SSPGRRPRGD VESRLDALQR QLNRLETRLS ADMATVLQ L LQRQMTLVPP AYSAVTTPGP GPTSTSPLLP VSPLPTLTLD SLSQVSQFMA CEELPPGAPE LPQEGPTRRL SLPGQLGAL TSQPLHRHGS DPGSLEVLFQ |
-Macromolecule #2: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 2 / Number of copies: 3 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Details: The grid was washed by acetone prior to use. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 291 K / Instrument: FEI VITROBOT MARK IV / Details: Blotting time was 5 seconds (blot force 10). |
| Details | This sample was mono-disperse. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Number grids imaged: 1 / Number real images: 1865 / Average exposure time: 55.55 sec. / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 120000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7cn1: |
 Movie
Movie Controller
Controller











 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)