+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the IL-17A-IL-17RA-IL-17RC ternary complex | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Receptor complex / IL-17A / IL-17RA / IL-17RC / cytokine | |||||||||
| Function / homology |  Function and homology information Function and homology informationinterleukin-17 receptor activity / granulocyte chemotaxis / positive regulation of interleukin-16 production / granulocyte migration / positive regulation of antimicrobial peptide production / Interleukin-17 signaling / T-helper 17 type immune response / cell death / interleukin-17A-mediated signaling pathway / positive regulation of interleukin-23 production ...interleukin-17 receptor activity / granulocyte chemotaxis / positive regulation of interleukin-16 production / granulocyte migration / positive regulation of antimicrobial peptide production / Interleukin-17 signaling / T-helper 17 type immune response / cell death / interleukin-17A-mediated signaling pathway / positive regulation of interleukin-23 production / negative regulation of inflammatory response to wounding / positive regulation of chemokine (C-X-C motif) ligand 1 production / interleukin-17-mediated signaling pathway / intestinal epithelial structure maintenance / positive regulation of interleukin-5 production / positive regulation of interleukin-13 production / fibroblast activation / positive regulation of bicellular tight junction assembly / positive regulation of osteoclast differentiation / positive regulation of cytokine production involved in inflammatory response / mRNA stabilization / keratinocyte proliferation / cellular response to interleukin-1 / defense response to fungus / coreceptor activity / keratinocyte differentiation / Notch signaling pathway / positive regulation of interleukin-12 production / cytokine activity / positive regulation of interleukin-1 beta production / protein catabolic process / response to wounding / positive regulation of interleukin-6 production / response to virus / positive regulation of tumor necrosis factor production / positive regulation of inflammatory response / cell-cell signaling / Interleukin-4 and Interleukin-13 signaling / gene expression / defense response to Gram-negative bacterium / adaptive immune response / cell surface receptor signaling pathway / immune response / defense response to Gram-positive bacterium / inflammatory response / protein heterodimerization activity / signaling receptor binding / external side of plasma membrane / innate immune response / apoptotic process / SARS-CoV-2 activates/modulates innate and adaptive immune responses / cell surface / protein homodimerization activity / positive regulation of transcription by RNA polymerase II / extracellular space / extracellular region / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
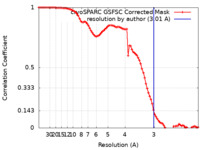
| Method | single particle reconstruction / cryo EM / Resolution: 3.01 Å | |||||||||
 Authors Authors | Wilson SC / Caveney NA / Jude KM / Garcia KC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Organizing structural principles of the IL-17 ligand-receptor axis. Authors: Steven C Wilson / Nathanael A Caveney / Michelle Yen / Christoph Pollmann / Xinyu Xiang / Kevin M Jude / Maximillian Hafer / Naotaka Tsutsumi / Jacob Piehler / K Christopher Garcia /   Abstract: The IL-17 family of cytokines and receptors have central roles in host defence against infection and development of inflammatory diseases. The compositions and structures of functional IL-17 family ...The IL-17 family of cytokines and receptors have central roles in host defence against infection and development of inflammatory diseases. The compositions and structures of functional IL-17 family ligand-receptor signalling assemblies remain unclear. IL-17E (also known as IL-25) is a key regulator of type 2 immune responses and driver of inflammatory diseases, such as allergic asthma, and requires both IL-17 receptor A (IL-17RA) and IL-17RB to elicit functional responses. Here we studied IL-25-IL-17RB binary and IL-25-IL-17RB-IL-17RA ternary complexes using a combination of cryo-electron microscopy, single-molecule imaging and cell-based signalling approaches. The IL-25-IL-17RB-IL-17RA ternary signalling assembly is a C2-symmetric complex in which the IL-25-IL-17RB homodimer is flanked by two 'wing-like' IL-17RA co-receptors through a 'tip-to-tip' geometry that is the key receptor-receptor interaction required for initiation of signal transduction. IL-25 interacts solely with IL-17RB to allosterically promote the formation of the IL-17RB-IL-17RA tip-to-tip interface. The resulting large separation between the receptors at the membrane-proximal level may reflect proximity constraints imposed by the intracellular domains for signalling. Cryo-electron microscopy structures of IL-17A-IL-17RA and IL-17A-IL-17RA-IL-17RC complexes reveal that this tip-to-tip architecture is a key organizing principle of the IL-17 receptor family. Furthermore, these studies reveal dual actions for IL-17RA sharing among IL-17 cytokine complexes, by either directly engaging IL-17 cytokines or alternatively functioning as a co-receptor. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26837.map.gz emd_26837.map.gz | 54.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26837-v30.xml emd-26837-v30.xml emd-26837.xml emd-26837.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26837_fsc.xml emd_26837_fsc.xml | 8.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_26837.png emd_26837.png | 53.5 KB | ||
| Masks |  emd_26837_msk_1.map emd_26837_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26837.cif.gz emd-26837.cif.gz | 7 KB | ||
| Others |  emd_26837_half_map_1.map.gz emd_26837_half_map_1.map.gz emd_26837_half_map_2.map.gz emd_26837_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26837 http://ftp.pdbj.org/pub/emdb/structures/EMD-26837 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26837 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26837 | HTTPS FTP |
-Related structure data
| Related structure data |  7uwnMC  7uwjC  7uwkC  7uwlC  7uwmC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26837.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26837.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.066 Å | ||||||||||||||||||||||||||||||||||||
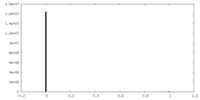
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26837_msk_1.map emd_26837_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
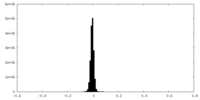
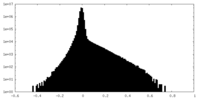
| Density Histograms |
-Half map: half map A
| File | emd_26837_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
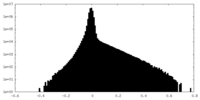
| Density Histograms |
-Half map: half map B
| File | emd_26837_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
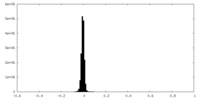
| Density Histograms |
- Sample components
Sample components
-Entire : IL-17A-IL-17RA-IL-17RC ternary complex
| Entire | Name: IL-17A-IL-17RA-IL-17RC ternary complex |
|---|---|
| Components |
|
-Supramolecule #1: IL-17A-IL-17RA-IL-17RC ternary complex
| Supramolecule | Name: IL-17A-IL-17RA-IL-17RC ternary complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Interleukin-17A
| Macromolecule | Name: Interleukin-17A / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 19.311678 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GITIPRNPGC PNSEDKNFPR TVMVNLNIHN RNTNTNPKRS SDYYNRSTSP WNLHRNEDPE RYPSVIWEAK CRHLGCINAD GNVDYHMNS VPIQQEILVL RREPPHCPNS FRLEKILVSV GCTCVTPIVH HVAGAPGSAL EVLFQGPGAA GLNDIFEAQK I EWHEHHHH HH UniProtKB: Interleukin-17A |
-Macromolecule #2: Interleukin-17 receptor A
| Macromolecule | Name: Interleukin-17 receptor A / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 36.888617 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LRLLDHRALV CSQPGLNCTV KNSTCLDDSW IHPRNLTPSS PKDLQIQLHF AHTQQGDLFP VAHIEWTLQT DASILYLEGA ELSVLQLNT NERLCVRFEF LSKLRHHHRR WRFTFSHFVV DPDQEYEVTV HHLPKPIPDG DPNHQSKNFL VPDCEHARMK V TTPCMSSG ...String: LRLLDHRALV CSQPGLNCTV KNSTCLDDSW IHPRNLTPSS PKDLQIQLHF AHTQQGDLFP VAHIEWTLQT DASILYLEGA ELSVLQLNT NERLCVRFEF LSKLRHHHRR WRFTFSHFVV DPDQEYEVTV HHLPKPIPDG DPNHQSKNFL VPDCEHARMK V TTPCMSSG SLWDPNITVE TLEAHQLRVS FTLWNESTHY QILLTSFPHM ENHSCFEHMH HIPAPRPEEF HQRSNVTLTL RN LKGCCRH QVQIQPFFSS CLNDCLRHSA TVSCPEMPDT PEPIPDYMSA ALEVLFQGPG AAEDQVDPRL IDGKHHHHHH HH UniProtKB: Interleukin-17 receptor A |
-Macromolecule #3: Isoform 5 of Interleukin-17 receptor C
| Macromolecule | Name: Isoform 5 of Interleukin-17 receptor C / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 53.294309 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LERLVGPQDA THCSPGLSCR LWDSDILCLP GDIVPAPGPV LAPTHLQTEL VLRCQKETDC DLCLRVAVHL AVHGHWEEPE DEEKFGGAA DLGVEEPRNA SLQAQVVLSF QAYPTARCVL LEVQVPAALV QFGQSVGSVV YDCFEAALGS EVRIWSYTQP R YEKELNHT ...String: LERLVGPQDA THCSPGLSCR LWDSDILCLP GDIVPAPGPV LAPTHLQTEL VLRCQKETDC DLCLRVAVHL AVHGHWEEPE DEEKFGGAA DLGVEEPRNA SLQAQVVLSF QAYPTARCVL LEVQVPAALV QFGQSVGSVV YDCFEAALGS EVRIWSYTQP R YEKELNHT QQLPDCRGLE VWNSIPSCWA LPWLNVSADG DNVHLVLNVS EEQHFGLSLY WNQVQGPPKP RWHKNLTGPQ II TLNHTDL VPCLCIQVWP LEPDSVRTNI CPFREDPRAH QNLWQAARLR LLTLQSWLLD APCSLPAEAA LCWRAPGGDP CQP LVPPLS WENVTVDKVL EFPLLKGHPN LCVQVNSSEK LQLQECLWAD SLGPLKDDVL LLETRGPQDN RSLCALEPSG CTSL PSKAS TRAARLGEYL LQDLQSGQCL QLWDDDLGAL WACPMDKYIH AAALEVLFQG PGAAEDQVDP RLIDGKHHHH HHHH UniProtKB: Interleukin-17 receptor C |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 9 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)