+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the IL-25-IL-17RB-IL-17RA ternary complex | |||||||||
 Map data Map data | Full map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Receptor complex / IL-17E / IL-25 / IL-17RB / IL-17RA / cytokine | |||||||||
| Function / homology |  Function and homology information Function and homology informationinterleukin-17E receptor binding / interleukin-17 receptor activity / eosinophil differentiation / granulocyte chemotaxis / Interleukin-17 signaling / T-helper 17 type immune response / interleukin-17A-mediated signaling pathway / positive regulation of interleukin-23 production / positive regulation of chemokine (C-X-C motif) ligand 1 production / response to fungus ...interleukin-17E receptor binding / interleukin-17 receptor activity / eosinophil differentiation / granulocyte chemotaxis / Interleukin-17 signaling / T-helper 17 type immune response / interleukin-17A-mediated signaling pathway / positive regulation of interleukin-23 production / positive regulation of chemokine (C-X-C motif) ligand 1 production / response to fungus / interleukin-17-mediated signaling pathway / positive regulation of interleukin-5 production / positive regulation of interleukin-13 production / fibroblast activation / response to nematode / inflammatory response to antigenic stimulus / positive regulation of cytokine production involved in inflammatory response / cytokine receptor activity / defense response to fungus / cytokine activity / protein catabolic process / regulation of cell growth / defense response / positive regulation of interleukin-6 production / response to virus / positive regulation of inflammatory response / cell surface receptor signaling pathway / inflammatory response / intracellular membrane-bounded organelle / signaling receptor binding / innate immune response / SARS-CoV-2 activates/modulates innate and adaptive immune responses / cell surface / positive regulation of transcription by RNA polymerase II / extracellular space / extracellular region / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
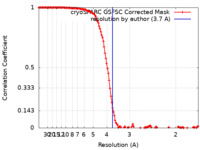
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Wilson SC / Caveney NA / Jude KM / Garcia KC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2022 Journal: Nature / Year: 2022Title: Organizing structural principles of the IL-17 ligand-receptor axis. Authors: Steven C Wilson / Nathanael A Caveney / Michelle Yen / Christoph Pollmann / Xinyu Xiang / Kevin M Jude / Maximillian Hafer / Naotaka Tsutsumi / Jacob Piehler / K Christopher Garcia /   Abstract: The IL-17 family of cytokines and receptors have central roles in host defence against infection and development of inflammatory diseases. The compositions and structures of functional IL-17 family ...The IL-17 family of cytokines and receptors have central roles in host defence against infection and development of inflammatory diseases. The compositions and structures of functional IL-17 family ligand-receptor signalling assemblies remain unclear. IL-17E (also known as IL-25) is a key regulator of type 2 immune responses and driver of inflammatory diseases, such as allergic asthma, and requires both IL-17 receptor A (IL-17RA) and IL-17RB to elicit functional responses. Here we studied IL-25-IL-17RB binary and IL-25-IL-17RB-IL-17RA ternary complexes using a combination of cryo-electron microscopy, single-molecule imaging and cell-based signalling approaches. The IL-25-IL-17RB-IL-17RA ternary signalling assembly is a C2-symmetric complex in which the IL-25-IL-17RB homodimer is flanked by two 'wing-like' IL-17RA co-receptors through a 'tip-to-tip' geometry that is the key receptor-receptor interaction required for initiation of signal transduction. IL-25 interacts solely with IL-17RB to allosterically promote the formation of the IL-17RB-IL-17RA tip-to-tip interface. The resulting large separation between the receptors at the membrane-proximal level may reflect proximity constraints imposed by the intracellular domains for signalling. Cryo-electron microscopy structures of IL-17A-IL-17RA and IL-17A-IL-17RA-IL-17RC complexes reveal that this tip-to-tip architecture is a key organizing principle of the IL-17 receptor family. Furthermore, these studies reveal dual actions for IL-17RA sharing among IL-17 cytokine complexes, by either directly engaging IL-17 cytokines or alternatively functioning as a co-receptor. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26835.map.gz emd_26835.map.gz | 180.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26835-v30.xml emd-26835-v30.xml emd-26835.xml emd-26835.xml | 21.8 KB 21.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_26835_fsc.xml emd_26835_fsc.xml | 14.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_26835.png emd_26835.png | 56.5 KB | ||
| Masks |  emd_26835_msk_1.map emd_26835_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-26835.cif.gz emd-26835.cif.gz | 6.9 KB | ||
| Others |  emd_26835_half_map_1.map.gz emd_26835_half_map_1.map.gz emd_26835_half_map_2.map.gz emd_26835_half_map_2.map.gz | 200 MB 200 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26835 http://ftp.pdbj.org/pub/emdb/structures/EMD-26835 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26835 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26835 | HTTPS FTP |
-Related structure data
| Related structure data |  7uwlMC  7uwjC  7uwkC  7uwmC  7uwnC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_26835.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26835.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Full map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8521 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_26835_msk_1.map emd_26835_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
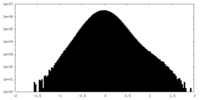
| Density Histograms |
-Half map: Half map B
| File | emd_26835_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map A
| File | emd_26835_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ternary IL-25-IL-17RB-IL-17RA complex
| Entire | Name: ternary IL-25-IL-17RB-IL-17RA complex |
|---|---|
| Components |
|
-Supramolecule #1: ternary IL-25-IL-17RB-IL-17RA complex
| Supramolecule | Name: ternary IL-25-IL-17RB-IL-17RA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Interleukin-25
| Macromolecule | Name: Interleukin-25 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 21.490201 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DASATHTYSH WPSCCPSKGQ DTSEELLRWS TVPVPPLEPA RPNRHPESCR ASEDGPLNSR AISPWRYELD RDLNRLPQDL YHARCLCPH CVSLQTGSHM DPRGNSELLY HNQTVFYRRP CHGEKGTHKG YCLERRLYRV SLACVCVRPR VMGAPAALEV L FQGPGAAG LNDIFEAQKI EWHEHHHHHH UniProtKB: Interleukin-25 |
-Macromolecule #2: Interleukin-17 receptor B
| Macromolecule | Name: Interleukin-17 receptor B / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 33.649098 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: REPTVQCGSE TGPSPEWMLQ HDLIPGDLRD LRVEPVTTSV ATGDYSILMN VSWVLRADAS IRLLKATKIC VTGKSNFQSY SCVRCNYTE AFQTQTRPSG GKWTFSYIGF PVELNTVYFI GAHNIPNANM NEDGPSMSVN FTSPGCLDHI MKYKKKCVKA G SLWDPNIT ...String: REPTVQCGSE TGPSPEWMLQ HDLIPGDLRD LRVEPVTTSV ATGDYSILMN VSWVLRADAS IRLLKATKIC VTGKSNFQSY SCVRCNYTE AFQTQTRPSG GKWTFSYIGF PVELNTVYFI GAHNIPNANM NEDGPSMSVN FTSPGCLDHI MKYKKKCVKA G SLWDPNIT ACKKNEETVE VNFTTTPLGN RYMALIQHST IIGFSQVFEP HQKKQTRASV VIPVTGDSEG ATVQLTPYFP TC GSDCIRH KGTVVLCPQT GVPFPLDNNK SKPGAAALEV LFQGPGAAED QVDPRLIDGK HHHHHHHH UniProtKB: Interleukin-17 receptor B |
-Macromolecule #3: Interleukin-17 receptor A
| Macromolecule | Name: Interleukin-17 receptor A / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 36.888617 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: LRLLDHRALV CSQPGLNCTV KNSTCLDDSW IHPRNLTPSS PKDLQIQLHF AHTQQGDLFP VAHIEWTLQT DASILYLEGA ELSVLQLNT NERLCVRFEF LSKLRHHHRR WRFTFSHFVV DPDQEYEVTV HHLPKPIPDG DPNHQSKNFL VPDCEHARMK V TTPCMSSG ...String: LRLLDHRALV CSQPGLNCTV KNSTCLDDSW IHPRNLTPSS PKDLQIQLHF AHTQQGDLFP VAHIEWTLQT DASILYLEGA ELSVLQLNT NERLCVRFEF LSKLRHHHRR WRFTFSHFVV DPDQEYEVTV HHLPKPIPDG DPNHQSKNFL VPDCEHARMK V TTPCMSSG SLWDPNITVE TLEAHQLRVS FTLWNESTHY QILLTSFPHM ENHSCFEHMH HIPAPRPEEF HQRSNVTLTL RN LKGCCRH QVQIQPFFSS CLNDCLRHSA TVSCPEMPDT PEPIPDYMSA ALEVLFQGPG AAEDQVDPRL IDGKHHHHHH HH UniProtKB: Interleukin-17 receptor A |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 14 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 53.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)