[English] 日本語
 Yorodumi
Yorodumi- EMDB-26748: Cryogenic electron microscopy 3D map of alpha-catenin ortholog, HMP1 -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryogenic electron microscopy 3D map of alpha-catenin ortholog, HMP1 | |||||||||
 Map data Map data | Cryo-EM 3D map of HMP1 | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 7.8 Å | |||||||||
 Authors Authors | Smith EW / Izard T / Rangarajan ES | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: J Biol Chem / Year: 2023 Journal: J Biol Chem / Year: 2023Title: The nematode α-catenin ortholog, HMP1, has an extended α-helix when bound to actin filaments. Authors: Erumbi S Rangarajan / Emmanuel W Smith / Tina Izard /  Abstract: The regulation of cell-cell junctions during epidermal morphogenesis ensures tissue integrity, a process regulated by α-catenin. This cytoskeletal protein connects the cadherin complex to ...The regulation of cell-cell junctions during epidermal morphogenesis ensures tissue integrity, a process regulated by α-catenin. This cytoskeletal protein connects the cadherin complex to filamentous actin at cell-cell junctions. The cadherin-catenin complex plays key roles in cell physiology, organism development, and disease. While mutagenesis of Caenorhabditis elegans cadherin and catenin shows that these proteins are key for embryonic morphogenesis, we know surprisingly little about their structure and attachment to the cytoskeleton. In contrast to mammalian α-catenin that functions as a dimer or monomer, the α-catenin ortholog from C. elegans, HMP1 for humpback, is a monomer. Our cryogenic electron microscopy (cryoEM) structure of HMP1/α-catenin reveals that the amino- and carboxy-terminal domains of HMP1/α-catenin are disordered and not in contact with the remaining HMP1/α-catenin middle domain. Since the carboxy-terminal HMP1/α-catenin domain is the F-actin-binding domain (FABD), this interdomain constellation suggests that HMP1/α-catenin is constitutively active, which we confirm biochemically. Our perhaps most surprising finding, given the high sequence similarity between the mammalian and nematode proteins, is our cryoEM structure of HMP1/α-catenin bound to F-actin. Unlike the structure of mammalian α-catenin bound to F-actin, binding to F-actin seems to allosterically convert a loop region of the HMP1/α-catenin FABD to extend an HMP1/α-catenin FABD α-helix. We use cryoEM and bundling assays to show for the first time how the FABD of HMP1/α-catenin bundles actin in the absence of force. Collectively, our data advance our understanding of α-catenin regulation of cell-cell contacts and additionally aid our understanding of the evolution of multicellularity in metazoans. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_26748.map.gz emd_26748.map.gz | 12.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-26748-v30.xml emd-26748-v30.xml emd-26748.xml emd-26748.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
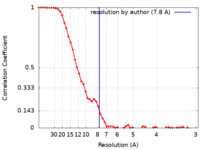
| FSC (resolution estimation) |  emd_26748_fsc.xml emd_26748_fsc.xml | 7 KB | Display |  FSC data file FSC data file |
| Images |  emd_26748.png emd_26748.png | 48.5 KB | ||
| Others |  emd_26748_half_map_1.map.gz emd_26748_half_map_1.map.gz emd_26748_half_map_2.map.gz emd_26748_half_map_2.map.gz | 12 MB 12 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-26748 http://ftp.pdbj.org/pub/emdb/structures/EMD-26748 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26748 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-26748 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_26748.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_26748.map.gz / Format: CCP4 / Size: 12.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM 3D map of HMP1 | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.48 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map A
| File | emd_26748_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
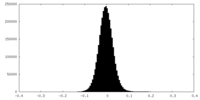
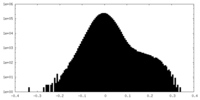
| Density Histograms |
-Half map: half map B
| File | emd_26748_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
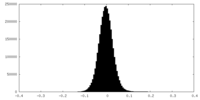
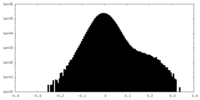
| Density Histograms |
- Sample components
Sample components
-Entire : alpha-catenin ortholog, HMP1
| Entire | Name: alpha-catenin ortholog, HMP1 |
|---|---|
| Components |
|
-Supramolecule #1: alpha-catenin ortholog, HMP1
| Supramolecule | Name: alpha-catenin ortholog, HMP1 / type: complex / ID: 1 / Chimera: Yes / Parent: 0 / Macromolecule list: all / Details: Full-length alpha-catenin ortholog, HMP1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 103.99428 KDa |
-Macromolecule #1: alpha-catenin ortholog, HMP1
| Macromolecule | Name: alpha-catenin ortholog, HMP1 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Sequence | String: MPANGNSHAY FNIDEVRSKN VLKQITQLIN EVTNITETFP LKPGQTTEGL VATLDAAVAN FLQTGSFAI SKCPIANSDP RAIDLLHEAL GAVQDTGQVM IQTGRDFVRD STSTNKRAIA T NSGRNLLT AVAKFLILAD SIDVKVIVDK VDEVRETAHQ MIEADTKIKV ...String: MPANGNSHAY FNIDEVRSKN VLKQITQLIN EVTNITETFP LKPGQTTEGL VATLDAAVAN FLQTGSFAI SKCPIANSDP RAIDLLHEAL GAVQDTGQVM IQTGRDFVRD STSTNKRAIA T NSGRNLLT AVAKFLILAD SIDVKVIVDK VDEVRETAHQ MIEADTKIKV DDLYNLLISQ IE ELDITVR RRAIDLVKPN QRDDLLAARS ALRQTAPLLY TSTRTFVRHP EHEEARRNRD YTA DEMHSA LNALESVLNG QQPKVTFSEY GRIGDLINEI DTFQNRIEID PAHYRRGTDR PDLE GHCER IVSGSASIAD AESTRENRKQ KIVAECNNLR QALQELLTEY EKSTGRRDDN DDIPL GIAE VHKRTKDLRR HLRRAIVDHI SDAFLDTRTP LILLIEAAKE GHEENTRYRS KMFQEH ANE IVSVARLSCQ LSSDVESVSV IQHTAAQLEK LAPQVAQAAI LLCHQPTSKT AQENMET YK NAWFDKVRLL TTALDNITTL DDFLAVSEAH IVEDCERGIK GITANASTPD ENAANCET V DCAAGSIRGR ALRVCDVVDA EMDFLQNSEY TETVKQAVRI LKTQRVDQFA ERASALANR QEAHGLTWDP KTKEEEMNEF INACTLVHDA VKDIRHALLM NRSMNDVDSD VEYEADGVGA ANADANRTI SEQENQQNLM RRLPEEEKKK IQAQIDIFKV TQTRFEREVA KWDETGNDII S LANNMCKI MMSMTEFTRG CGPLKTTMDV IRAAQEISLN GSKLNALARQ IGEESADSQT KK DLLAYLS QITLYCQQLN ICSKVKADVT QVGNELVVSA LDSAMSLIQT ARNLLTAVVQ TVK AAYIAS TKFRRPNANS VRVEWRMAPP KKQPLIRPQK NNAIIRRASE RRPLQPAKVL AEFT RNEIE TGRDSDDEEL DRRHQQRING RL |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 240 sec. | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 277 K / Instrument: LEICA EM GP |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 1294 / Average exposure time: 0.1 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: JEOL CRYOSPECPORTER / Cooling holder cryogen: NITROGEN |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Protocol: FLEXIBLE FIT |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)