[English] 日本語
 Yorodumi
Yorodumi- EMDB-25694: Cryo-EM structure of ATP-bound PCAT1 in the outward-facing confor... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-25694 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of ATP-bound PCAT1 in the outward-facing conformation | |||||||||
 Map data Map data | full map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ATP-binding cassette / ABC transporter / protein transport / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationABC-type bacteriocin transporter activity / ABC-type oligopeptide transporter activity / cysteine-type peptidase activity / ATP hydrolysis activity / proteolysis / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Acetivibrio thermocellus (bacteria) Acetivibrio thermocellus (bacteria) | |||||||||
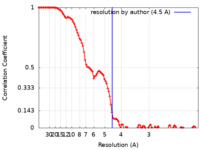
| Method | single particle reconstruction / cryo EM / Resolution: 4.5 Å | |||||||||
 Authors Authors | Kieuvongngam V / Chen J | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2022 Journal: Proc Natl Acad Sci U S A / Year: 2022Title: Structures of the peptidase-containing ABC transporter PCAT1 under equilibrium and nonequilibrium conditions. Authors: Virapat Kieuvongngam / Jue Chen /  Abstract: ATP-binding cassette (ABC) transporters are ubiquitous molecular pumps that transport a broad range of substrates across biological membranes. Although the structure and function of ABC transporters ...ATP-binding cassette (ABC) transporters are ubiquitous molecular pumps that transport a broad range of substrates across biological membranes. Although the structure and function of ABC transporters has been studied extensively, our understanding of their energetics and dynamics remains limited. Here, we present studies of the peptidase-containing ABC transporter 1 (PCAT1), a polypeptide processing and secretion ABC transporter that functions via the classic alternating access mechanism. PCAT1 is a homodimer containing two peptidase (PEP) domains, two transmembrane domains, and two nucleotide-binding domains (NBDs). Using cryo-electron microscopy, we analyzed the structures of wild-type PCAT1 under conditions that either prevent or permit ATP hydrolysis and observed two completely different conformational distributions. In the presence of ATP but absence of Mg, PCAT1 adopts an NBD-dimerized, outward-facing conformation. The two PEP domains are dissociated from the transporter core, preventing uncoupled substrate cleavage. The addition of Mg to promote ATP hydrolysis shifts the majority of the particles into NBD-separated, inward-facing conformations. Under this ATP turnover condition, only a small fraction of PCAT1 adopts the NBD-dimerized conformation. These data give rise to two mechanistic conclusions: 1) the ATP-bound, NBD-dimerized conformation is the lowest energy state, and 2) the rate-limiting step in the PCAT1 transport cycle is the formation of the NBD dimer. The thermodynamic conclusion is likely a general property shared by many ABC transporters. The kinetic bottleneck, however, varies from transporter to transporter. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_25694.map.gz emd_25694.map.gz | 50.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-25694-v30.xml emd-25694-v30.xml emd-25694.xml emd-25694.xml | 22.6 KB 22.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_25694_fsc.xml emd_25694_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_25694.png emd_25694.png | 42 KB | ||
| Filedesc metadata |  emd-25694.cif.gz emd-25694.cif.gz | 7.1 KB | ||
| Others |  emd_25694_half_map_1.map.gz emd_25694_half_map_1.map.gz emd_25694_half_map_2.map.gz emd_25694_half_map_2.map.gz | 95.4 MB 95.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-25694 http://ftp.pdbj.org/pub/emdb/structures/EMD-25694 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25694 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-25694 | HTTPS FTP |
-Related structure data
| Related structure data |  7t54MC  7t55C  7t56C  7t57C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-11009 (Title: Cryo-electron microscopy reconstruction of ATP-bound PCAT1 in an outward-facing conformation in a Mg2+ free condition EMPIAR-11009 (Title: Cryo-electron microscopy reconstruction of ATP-bound PCAT1 in an outward-facing conformation in a Mg2+ free conditionData size: 2.6 TB Data #1: Unaligned and uncorrected multiframe movies of ATP-bound PCAT1 in an outward-facing conformation from a Mg2+ free condition [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_25694.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_25694.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.03 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: half map A
| File | emd_25694_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map B
| File | emd_25694_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : PCAT1 homodimer
| Entire | Name: PCAT1 homodimer |
|---|---|
| Components |
|
-Supramolecule #1: PCAT1 homodimer
| Supramolecule | Name: PCAT1 homodimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Acetivibrio thermocellus (bacteria) Acetivibrio thermocellus (bacteria) |
| Molecular weight | Theoretical: 162.176 KDa |
-Macromolecule #1: ABC-type bacteriocin transporter
| Macromolecule | Name: ABC-type bacteriocin transporter / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acetivibrio thermocellus (bacteria) Acetivibrio thermocellus (bacteria)Strain: ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372 |
| Molecular weight | Theoretical: 81.180805 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNAMLRRLFK KKYVCVRQYD LTDCGAACLS SIAQYYGLKM SLAKIREMTG TDTQGTNAYG LIHAAKQLGF SAKGVKASKE DLLKDFRLP AIANVIVDNR LAHFVVIYSI KNRIITVADP GKGIVRYSMD DFCSIWTGGL VLLEPGEAFQ KGDYTQNMMV K FAGFLKPL ...String: SNAMLRRLFK KKYVCVRQYD LTDCGAACLS SIAQYYGLKM SLAKIREMTG TDTQGTNAYG LIHAAKQLGF SAKGVKASKE DLLKDFRLP AIANVIVDNR LAHFVVIYSI KNRIITVADP GKGIVRYSMD DFCSIWTGGL VLLEPGEAFQ KGDYTQNMMV K FAGFLKPL KKTVLCIFLA SLLYTALGIA GSFYIKFLFD DLIKFEKLND LHIISAGFAV IFLLQIFLNY YRSILVTKLG MS IDKSIMM EYYSHVLKLP MNFFNSRKVG EIISRFMDAS KIRQAISGAT LTIMIDTIMA VIGGILLYIQ NSSLFFISFI IIL LYGIIV TVFNKPIQNA NRQIMEDNAK LTSALVESVK GIETIKSFGA EEQTEKSTRD KIETVMKSSF KEGMLYINLS SLTG IVAGL GGIVILWAGA YNVIKGNMSG GQLLAFNALL AYFLTPVKNL IDLQPLIQTA VVASNRLGEI LELATEKELR EDSDD FVIS LKGDIEFRNV DFRYGLRKPV LKNINLTIPK GKTVAIVGES GSGKTTLAKL LMNFYSPEKG DILINGHSIK NISLEL IRK KIAFVSQDVF IFSGTVKENL CLGNENVDMD EIIKAAKMAN AHDFIEKLPL KYDTFLNESG ANLSEGQKQR LAIARAL LK KPDILILDEA TSNLDSITEN HIKDAIYGLE DDVTVIIIAH RLSTIVNCDK IYLLKDGEIV ESGSHTELIA LKGCYFKM W KQTENTLAS UniProtKB: ABC-type bacteriocin transporter |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 2 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 5 mg/mL | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| ||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 20 sec. / Pretreatment - Atmosphere: AIR | ||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 100.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-50 / Number grids imaged: 2 / Number real images: 5994 / Average exposure time: 0.2 sec. / Average electron dose: 1.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.8 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)