+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23908 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | RNA polymerase II initially transcribing complex (ITC) | ||||||||||||||||||
 Map data Map data | ITC | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | PIC / TFIIH / transcription / ITC / RNA polymerase II | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology information: / : / : / : / regulation of mitotic recombination / RNA polymerase II complex recruiting activity / transcription open complex formation at RNA polymerase II promoter / TFIIA-class transcription factor complex binding / RNA polymerase III transcription regulatory region sequence-specific DNA binding / : ...: / : / : / : / regulation of mitotic recombination / RNA polymerase II complex recruiting activity / transcription open complex formation at RNA polymerase II promoter / TFIIA-class transcription factor complex binding / RNA polymerase III transcription regulatory region sequence-specific DNA binding / : / TFIIH-class transcription factor complex binding / RNA polymerase III preinitiation complex assembly / RNA polymerase II promoter clearance / transcription factor TFIIIB complex / positive regulation of mitotic recombination / RNA polymerase I general transcription initiation factor binding / transcription factor TFIIE complex / nucleotide-excision repair factor 3 complex / nucleotide-excision repair, preincision complex assembly / DNA translocase activity / regulation of transcription by RNA polymerase III / TFIIF-class transcription factor complex binding / transcriptional start site selection at RNA polymerase II promoter / transcription factor TFIIF complex / hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides / transcription factor TFIIA complex / RNA polymerase I preinitiation complex assembly / transcription factor TFIIH core complex / transcription factor TFIIH holo complex / cyclin-dependent protein serine/threonine kinase activator activity / transcription preinitiation complex / Processing of Capped Intron-Containing Pre-mRNA / RNA Polymerase III Transcription Initiation From Type 2 Promoter / DNA binding, bending / RNA Pol II CTD phosphorylation and interaction with CE / Formation of the Early Elongation Complex / mRNA Capping / RNA polymerase II transcribes snRNA genes / TP53 Regulates Transcription of DNA Repair Genes / RNA Polymerase II Promoter Escape / RNA Polymerase II Transcription Pre-Initiation And Promoter Opening / RNA Polymerase II Transcription Initiation / RNA Polymerase II Transcription Initiation And Promoter Clearance / poly(A)+ mRNA export from nucleus / RNA polymerase II general transcription initiation factor activity / RNA Polymerase II Pre-transcription Events / transcription factor TFIID complex / Formation of TC-NER Pre-Incision Complex / positive regulation of nuclear-transcribed mRNA poly(A) tail shortening / 3'-5' DNA helicase activity / RNA Polymerase I Promoter Escape / DNA 3'-5' helicase / Gap-filling DNA repair synthesis and ligation in TC-NER / nucleolar large rRNA transcription by RNA polymerase I / Estrogen-dependent gene expression / RNA polymerase II complex binding / ATPase activator activity / maintenance of transcriptional fidelity during transcription elongation by RNA polymerase II / positive regulation of translational initiation / Dual incision in TC-NER / protein phosphatase activator activity / RNA polymerase I complex / RNA polymerase III complex / RNA polymerase II core promoter sequence-specific DNA binding / RNA polymerase II preinitiation complex assembly / RNA polymerase II, core complex / tRNA transcription by RNA polymerase III / transcription-coupled nucleotide-excision repair / translation initiation factor binding / TBP-class protein binding / DNA helicase activity / nucleotide-excision repair / transcription initiation at RNA polymerase II promoter / transcription elongation by RNA polymerase II / P-body / DNA-templated transcription initiation / positive regulation of transcription elongation by RNA polymerase II / ribonucleoside binding / DNA-directed RNA polymerase / disordered domain specific binding / DNA-directed RNA polymerase activity / single-stranded DNA binding / double-stranded DNA binding / transcription regulator complex / DNA-binding transcription factor binding / nucleic acid binding / transcription by RNA polymerase II / damaged DNA binding / DNA helicase / RNA polymerase II-specific DNA-binding transcription factor binding / protein dimerization activity / single-stranded RNA binding / nucleotide binding / negative regulation of DNA-templated transcription / chromatin binding / regulation of transcription by RNA polymerase II / regulation of DNA-templated transcription / DNA-templated transcription / nucleolus / positive regulation of transcription by RNA polymerase II Similarity search - Function | ||||||||||||||||||
| Biological species |  | ||||||||||||||||||
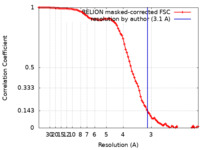
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | ||||||||||||||||||
 Authors Authors | Yang C / Fujiwara R | ||||||||||||||||||
| Funding support |  United States, 5 items United States, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2022 Journal: Mol Cell / Year: 2022Title: Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II. Authors: Chun Yang / Rina Fujiwara / Hee Jong Kim / Pratik Basnet / Yunye Zhu / Jose J Gorbea Colón / Stefan Steimle / Benjamin A Garcia / Craig D Kaplan / Kenji Murakami /  Abstract: Previous structural studies of the initiation-elongation transition of RNA polymerase II (pol II) transcription have relied on the use of synthetic oligonucleotides, often artificially discontinuous ...Previous structural studies of the initiation-elongation transition of RNA polymerase II (pol II) transcription have relied on the use of synthetic oligonucleotides, often artificially discontinuous to capture pol II in the initiating state. Here, we report multiple structures of initiation complexes converted de novo from a 33-subunit yeast pre-initiation complex (PIC) through catalytic activities and subsequently stalled at different template positions. We determine that PICs in the initially transcribing complex (ITC) can synthesize a transcript of ∼26 nucleotides before transitioning to an elongation complex (EC) as determined by the loss of general transcription factors (GTFs). Unexpectedly, transition to an EC was greatly accelerated when an ITC encountered a downstream EC stalled at promoter proximal regions and resulted in a collided head-to-end dimeric EC complex. Our structural analysis reveals a dynamic state of TFIIH, the largest of GTFs, in PIC/ITC with distinct functional consequences at multiple steps on the pathway to elongation. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23908.map.gz emd_23908.map.gz | 93 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23908-v30.xml emd-23908-v30.xml emd-23908.xml emd-23908.xml | 56.5 KB 56.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23908_fsc.xml emd_23908_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_23908.png emd_23908.png | 49.1 KB | ||
| Filedesc metadata |  emd-23908.cif.gz emd-23908.cif.gz | 14.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23908 http://ftp.pdbj.org/pub/emdb/structures/EMD-23908 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23908 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23908 | HTTPS FTP |
-Related structure data
| Related structure data |  7ml4MC  7meiC  7mk9C  7mkaC  7ml0C  7ml1C  7ml2C  7ml3C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10865 (Title: Structural visualization of de novo initiation of RNA polymerase II transcription EMPIAR-10865 (Title: Structural visualization of de novo initiation of RNA polymerase II transcriptionData size: 14.3 TB Data #1: raw micrographs for PIC + ITC maps [micrographs - multiframe] Data #2: raw micrographs for EC+EC map [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23908.map.gz / Format: CCP4 / Size: 364.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23908.map.gz / Format: CCP4 / Size: 364.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | ITC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : RNA polymerase II initially transcribing complex (ITC)
+Supramolecule #1: RNA polymerase II initially transcribing complex (ITC)
+Supramolecule #2: Pol II + TFIIB + RNA
+Supramolecule #3: DNA+TBP+TFIIA+TFIIE+TFIIF
+Supramolecule #4: TFIIH
+Macromolecule #1: Transcription initiation factor IIF subunit alpha
+Macromolecule #2: Transcription initiation factor IIF subunit beta
+Macromolecule #3: DNA-directed RNA polymerase II subunit RPB4
+Macromolecule #4: DNA-directed RNA polymerase II subunit RPB7
+Macromolecule #5: Transcription initiation factor IIB
+Macromolecule #6: DNA-directed RNA polymerase subunit
+Macromolecule #7: DNA-directed RNA polymerase subunit beta
+Macromolecule #8: DNA-directed RNA polymerase II subunit RPB3
+Macromolecule #9: DNA-directed RNA polymerases I, II, and III subunit RPABC1
+Macromolecule #10: DNA-directed RNA polymerases I,II,and III subunit RPABC2
+Macromolecule #11: DNA-directed RNA polymerases I, II, and III subunit RPABC3
+Macromolecule #12: DNA-directed RNA polymerase II subunit RPB9
+Macromolecule #13: DNA-directed RNA polymerases II subunit RPABC5
+Macromolecule #14: DNA-directed RNA polymerase II subunit RPB11
+Macromolecule #15: DNA-directed RNA polymerases II subunit RPABC4
+Macromolecule #16: BJ4_G0050160.mRNA.1.CDS.1
+Macromolecule #17: General transcription and DNA repair factor IIH helicase subunit XPD
+Macromolecule #18: General transcription and DNA repair factor IIH subunit TFB4
+Macromolecule #19: General transcription and DNA repair factor IIH
+Macromolecule #20: Tfb1
+Macromolecule #21: General transcription and DNA repair factor IIH helicase subunit XPB
+Macromolecule #22: General transcription and DNA repair factor IIH subunit TFB5
+Macromolecule #23: RNA polymerase II transcription factor B subunit 2
+Macromolecule #24: Transcription initiation factor IIE subunit beta
+Macromolecule #25: Transcription initiation factor IIA large subunit
+Macromolecule #26: Transcription initiation factor IIA subunit 2
+Macromolecule #29: TATA-box-binding protein
+Macromolecule #30: Transcription initiation factor IIE subunit alpha
+Macromolecule #27: non-template strand DNA
+Macromolecule #28: template strand DNA
+Macromolecule #31: RNA
+Macromolecule #32: ZINC ION
+Macromolecule #33: MAGNESIUM ION
+Macromolecule #34: IRON/SULFUR CLUSTER
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)