[English] 日本語
 Yorodumi
Yorodumi- EMDB-23807: Structure of yeast Ubr1 in complex with Ubc2 and monoubiquitinate... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23807 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of yeast Ubr1 in complex with Ubc2 and monoubiquitinated N-degron | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ubiquitin E3 ligase / ubiquitination / Ubr1 / Ubc2 / Degron / N-end rule / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationMUB1-RAD6-UBR2 ubiquitin ligase complex / RAD6-UBR2 ubiquitin ligase complex / Rad6-Rad18 complex / regulation of dipeptide transport / UBR1-RAD6 ubiquitin ligase complex / sno(s)RNA transcription / HULC complex / proteasome regulatory particle binding / error-free postreplication DNA repair / meiotic DNA double-strand break formation ...MUB1-RAD6-UBR2 ubiquitin ligase complex / RAD6-UBR2 ubiquitin ligase complex / Rad6-Rad18 complex / regulation of dipeptide transport / UBR1-RAD6 ubiquitin ligase complex / sno(s)RNA transcription / HULC complex / proteasome regulatory particle binding / error-free postreplication DNA repair / meiotic DNA double-strand break formation / ubiquitin-dependent protein catabolic process via the N-end rule pathway / stress-induced homeostatically regulated protein degradation pathway / Synthesis of active ubiquitin: roles of E1 and E2 enzymes / mitochondria-associated ubiquitin-dependent protein catabolic process / cytoplasm protein quality control by the ubiquitin-proteasome system / E3 ubiquitin ligases ubiquitinate target proteins / telomere maintenance via recombination / proteasome regulatory particle, base subcomplex / symbiont entry into host cell via disruption of host cell glycocalyx / ribosome-associated ubiquitin-dependent protein catabolic process / error-free translesion synthesis / E2 ubiquitin-conjugating enzyme / sporulation resulting in formation of a cellular spore / symbiont entry into host cell via disruption of host cell envelope / virus tail / proteasome binding / ubiquitin conjugating enzyme activity / Antigen processing: Ubiquitination & Proteasome degradation / protein monoubiquitination / cellular response to unfolded protein / ubiquitin ligase complex / error-prone translesion synthesis / subtelomeric heterochromatin formation / ERAD pathway / mitotic G1 DNA damage checkpoint signaling / double-strand break repair via homologous recombination / DNA-templated transcription termination / RING-type E3 ubiquitin transferase / protein polyubiquitination / ubiquitin-protein transferase activity / ubiquitin protein ligase activity / ubiquitin-dependent protein catabolic process / proteasome-mediated ubiquitin-dependent protein catabolic process / transcription by RNA polymerase II / chromosome, telomeric region / protein ubiquitination / DNA repair / chromatin / zinc ion binding / ATP binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.67 Å | |||||||||
 Authors Authors | Pan M / Zheng Q | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nature / Year: 2021 Journal: Nature / Year: 2021Title: Structural insights into Ubr1-mediated N-degron polyubiquitination. Authors: Man Pan / Qingyun Zheng / Tian Wang / Lujun Liang / Junxiong Mao / Chong Zuo / Ruichao Ding / Huasong Ai / Yuan Xie / Dong Si / Yuanyuan Yu / Lei Liu / Minglei Zhao /   Abstract: The N-degron pathway targets proteins that bear a destabilizing residue at the N terminus for proteasome-dependent degradation. In yeast, Ubr1-a single-subunit E3 ligase-is responsible for the Arg/N- ...The N-degron pathway targets proteins that bear a destabilizing residue at the N terminus for proteasome-dependent degradation. In yeast, Ubr1-a single-subunit E3 ligase-is responsible for the Arg/N-degron pathway. How Ubr1 mediates the initiation of ubiquitination and the elongation of the ubiquitin chain in a linkage-specific manner through a single E2 ubiquitin-conjugating enzyme (Ubc2) remains unknown. Here we developed chemical strategies to mimic the reaction intermediates of the first and second ubiquitin transfer steps, and determined the cryo-electron microscopy structures of Ubr1 in complex with Ubc2, ubiquitin and two N-degron peptides, representing the initiation and elongation steps of ubiquitination. Key structural elements, including a Ubc2-binding region and an acceptor ubiquitin-binding loop on Ubr1, were identified and characterized. These structures provide mechanistic insights into the initiation and elongation of ubiquitination catalysed by Ubr1. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23807.map.gz emd_23807.map.gz | 49.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23807-v30.xml emd-23807-v30.xml emd-23807.xml emd-23807.xml | 24.1 KB 24.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_23807.png emd_23807.png | 167.9 KB | ||
| Masks |  emd_23807_msk_1.map emd_23807_msk_1.map | 64 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-23807.cif.gz emd-23807.cif.gz | 7.3 KB | ||
| Others |  emd_23807_additional_1.map.gz emd_23807_additional_1.map.gz emd_23807_half_map_1.map.gz emd_23807_half_map_1.map.gz emd_23807_half_map_2.map.gz emd_23807_half_map_2.map.gz | 59.8 MB 49.6 MB 49.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23807 http://ftp.pdbj.org/pub/emdb/structures/EMD-23807 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23807 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23807 | HTTPS FTP |
-Validation report
| Summary document |  emd_23807_validation.pdf.gz emd_23807_validation.pdf.gz | 966.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23807_full_validation.pdf.gz emd_23807_full_validation.pdf.gz | 965.9 KB | Display | |
| Data in XML |  emd_23807_validation.xml.gz emd_23807_validation.xml.gz | 12.2 KB | Display | |
| Data in CIF |  emd_23807_validation.cif.gz emd_23807_validation.cif.gz | 14.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23807 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23807 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23807 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23807 | HTTPS FTP |
-Related structure data
| Related structure data |  7meyMC  7mexC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10902 (Title: Single-particle cryoEM data of yeast Ubr1-Ubc2-Ub-Ub-N-degron complex (elongation) EMPIAR-10902 (Title: Single-particle cryoEM data of yeast Ubr1-Ubc2-Ub-Ub-N-degron complex (elongation)Data size: 2.9 TB Data #1: Unaligned movie data of yeast Ubr1-Ubc2-Ub-Ub-N-degron complex (elongation complex) [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_23807.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23807.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.063 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_23807_msk_1.map emd_23807_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
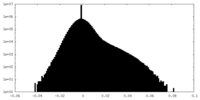
| Density Histograms |
-Additional map: #1
| File | emd_23807_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
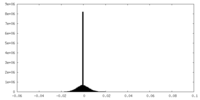
| Density Histograms |
-Half map: #2
| File | emd_23807_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_23807_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : yeast Ubr1 in complex with Ubc2 and monoubiquitinated N-degron
| Entire | Name: yeast Ubr1 in complex with Ubc2 and monoubiquitinated N-degron |
|---|---|
| Components |
|
-Supramolecule #1: yeast Ubr1 in complex with Ubc2 and monoubiquitinated N-degron
| Supramolecule | Name: yeast Ubr1 in complex with Ubc2 and monoubiquitinated N-degron type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: E3 ubiquitin-protein ligase UBR1
| Macromolecule | Name: E3 ubiquitin-protein ligase UBR1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RING-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 225.10275 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSVADDDLGS LQGHIRRTLR SIHNLPYFRY TRGPTERADM SRALKEFIYR YLYFVISNSG ENLPTLFNAH PKQKLSNPEL TVFPDSLED AVDIDKITSQ QTIPFYKIDE SRIGDVHKHT GRNCGRKFKI GEPLYRCHEC GCDDTCVLCI HCFNPKDHVN H HVCTDICT ...String: MSVADDDLGS LQGHIRRTLR SIHNLPYFRY TRGPTERADM SRALKEFIYR YLYFVISNSG ENLPTLFNAH PKQKLSNPEL TVFPDSLED AVDIDKITSQ QTIPFYKIDE SRIGDVHKHT GRNCGRKFKI GEPLYRCHEC GCDDTCVLCI HCFNPKDHVN H HVCTDICT EFTSGICDCG DEEAWNSPLH CKAEEQENDI SEDPATNADI KEEDVWNDSV NIALVELVLA EVFDYFIDVF NQ NIEPLPT IQKDITIKLR EMTQQGKMYE RAQFLNDLKY ENDYMFDGTT TAKTSPSNSP EASPSLAKID PENYTVIIYN DEY HNYSQA TTALRQGVPD NVHIDLLTSR IDGEGRAMLK CSQDLSSVLG GFFAVQTNGL SATLTSWSEY LHQETCKYII LWIT HCLNI PNSSFQTTFR NMMGKTLCSE YLNATECRDM TPVVEKYFSN KFDKNDPYRY IDLSILADGN QIPLGHHKIL PESST HSLS PLINDVETPT SRTYSNTRLQ HILYFDNRYW KRLRKDIQNV IIPTLASSNL YKPIFCQQVV EIFNHITRSV AYMDRE PQL TAIRECVVQL FTCPTNAKNI FENQSFLDIV WSIIDIFKEF CKVEGGVLIW QRVQKSNLTK SYSISFKQGL YTVETLL SK VHDPNIPLRP KEIISLLTLC KLFNGAWKIK RKEGEHVLHE DQNFISYLEY TTSIYSIIQT AEKVSEKSKD SIDSKLFL N AIRIISSFLG NRSLTYKLIY DSHEVIKFSV SHERVAFMNP LQTMLSFLIE KVSLKDAYEA LEDCSDFLKI SDFSLRSVV LCSQIDVGFW VRNGMSVLHQ ASYYKNNPEL GSYSRDIHLN QLAILWERDD IPRIIYNILD RWELLDWFTG EVDYQHTVYE DKISFIIQQ FIAFIYQILT ERQYFKTFSS LKDRRMDQIK NSIIYNLYMK PLSYSKLLRS VPDYLTEDTT EFDEALEEVS V FVEPKGLA DNGVFKLKAS LYAKVDPLKL LNLENEFESS ATIIKSHLAK DKDEIAKVVL IPQVSIKQLD KDALNLGAFT RN TVFAKVV YKLLQVCLDM EDSTFLNELL HLVHGIFRDD ELINGKDSIP EAYLSKPICN LLLSIANAKS DVFSESIVRK ADY LLEKMI MKKPNELFES LIASFGNQYV NDYKDKKLRQ GVNLQETEKE RKRRLAKKHQ ARLLAKFNNQ QTKFMKEHES EFDE QDNDV DMVGEKVYES EDFTCALCQD SSSTDFFVIP AYHDHSPIFR PGNIFNPNEF MPMWDGFYND DEKQAYIDDD VLEAL KENG SCGSRKVFVS CNHHIHHNCF KRYVQKKRFS SNAFICPLCQ TFSNCTLPLC QTSKANTGLS LDMFLESELS LDTLSR LFK PFTEENYRTI NSIFSLMISQ CQGFDKAVRK RANFSHKDVS LILSVHWANT ISMLEIASRL EKPYSISFFR SREQKYK TL KNILVCIMLF TFVIGKPSME FEPYPQQPDT VWNQNQLFQY IVRSALFSPV SLRQTVTEAL TTFSRQFLRD FLQGLSDA E QVTKLYAKAS KIGDVLKVSE QMLFALRTIS DVRMEGLDSE SIIYDLAYTF LLKSLLPTIR RCLVFIKVLH ELVKDSENE TLVINGHEVE EELEFEDTAE FVNKALKMIT EKESLVDLLT TQESIVSHPY LENIPYEYCG IIKLIDLSKY LNTYVTQSKE IKLREERSQ HMKNADNRLD FKICLTCGVK VHLRADRHEM TKHLNKNCFK PFGAFLMPNS SEVCLHLTQP PSNIFISAPY L NSHGEVGR NAMRRGDLTT LNLKRYEHLN RLWINNEIPG YISRVMGDEF RVTILSNGFL FAFNREPRPR RIPPTDEDDE DM EEGEDGF FTEGNDEMDV DDETGQAANL FGVGAEGIAG GGVRDFFQFF ENFRNTLQPQ GNGDDDAPQN PPPILQFLGP QFD GATIIR NTNPRNLDED DSDDNDDSDE REIW UniProtKB: E3 ubiquitin-protein ligase UBR1 |
-Macromolecule #2: Ubiquitin
| Macromolecule | Name: Ubiquitin / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8.493741 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MQIFVKTLTG KTITLEVEPS DTIENVKAKI QDKEGIPPDQ QRLIFAGCQL EDGRTLSDYN IQKESTLHLV LRLRG UniProtKB: Tail fiber |
-Macromolecule #3: Ubiquitin-conjugating enzyme E2 2
| Macromolecule | Name: Ubiquitin-conjugating enzyme E2 2 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: E2 ubiquitin-conjugating enzyme |
|---|---|
| Source (natural) | Organism:  Strain: ATCC 204508 / S288c |
| Molecular weight | Theoretical: 19.725662 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSTPARRRLM RDFKRMKEDA PPGVSASPLP DNVMVWNAMI IGPADTPYED GTFRLLLEFD EEYPNKPPHV KFLSEMFHPN VYANGEICL DILQNRWTPT YDVASILTSI QSLFNDPNPA SPANVEAATL FKDHKSQYVK RVKETVEKSW EDDMDDMDDD D DDDDDDDD DEAD UniProtKB: Ubiquitin-conjugating enzyme E2 2 |
-Macromolecule #4: Ubiquitin
| Macromolecule | Name: Ubiquitin / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8.519778 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MQIFVKTLTG KTITLEVEPS DTIENVKAKI QDKEGIPPDQ QRLIFAGKQL EDGRTLSDYN IQKESTLHLV LRLRG UniProtKB: Tail fiber |
-Macromolecule #5: Monoubiquitinated N-degron
| Macromolecule | Name: Monoubiquitinated N-degron / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 4.571223 KDa |
| Sequence | String: RHGSGSGAWL LPVSLVKRKT TLAPNTQTAS PPSYRALADS LMQ |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 7 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #7: 2-(ethylamino)ethane-1-thiol
| Macromolecule | Name: 2-(ethylamino)ethane-1-thiol / type: ligand / ID: 7 / Number of copies: 1 / Formula: Z3V |
|---|---|
| Molecular weight | Theoretical: 105.202 Da |
| Chemical component information |  ChemComp-Z3V: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 48.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-7mey: |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)