[English] 日本語
 Yorodumi
Yorodumi- EMDB-21921: Cryo-EM structure of Bacillus subtilis RNA Polymerase in complex ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21921 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Bacillus subtilis RNA Polymerase in complex with HelD | |||||||||
 Map data Map data | RNA Polymerase in complex with HelD | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA POLYMERASE / TRANSFERASE-HELD COMPLEX / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationnucleoid / recombinational repair / 3'-5' DNA helicase activity / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / DNA-directed RNA polymerase complex / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity ...nucleoid / recombinational repair / 3'-5' DNA helicase activity / bacterial-type RNA polymerase core enzyme binding / cytosolic DNA-directed RNA polymerase complex / DNA-directed RNA polymerase complex / DNA-templated transcription initiation / ribonucleoside binding / DNA-directed RNA polymerase / DNA-directed RNA polymerase activity / DNA helicase / protein dimerization activity / response to antibiotic / DNA-templated transcription / magnesium ion binding / ATP hydrolysis activity / DNA binding / zinc ion binding / ATP binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
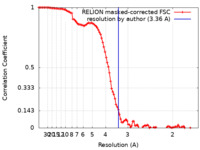
| Method | single particle reconstruction / cryo EM / Resolution: 3.36 Å | |||||||||
 Authors Authors | Newing T / Tolun G | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2020 Journal: Nat Commun / Year: 2020Title: Molecular basis for RNA polymerase-dependent transcription complex recycling by the helicase-like motor protein HelD. Authors: Timothy P Newing / Aaron J Oakley / Michael Miller / Catherine J Dawson / Simon H J Brown / James C Bouwer / Gökhan Tolun / Peter J Lewis /  Abstract: In bacteria, transcription complexes stalled on DNA represent a major source of roadblocks for the DNA replication machinery that must be removed in order to prevent damaging collisions. Gram- ...In bacteria, transcription complexes stalled on DNA represent a major source of roadblocks for the DNA replication machinery that must be removed in order to prevent damaging collisions. Gram-positive bacteria contain a transcription factor HelD that is able to remove and recycle stalled complexes, but it was not known how it performed this function. Here, using single particle cryo-electron microscopy, we have determined the structures of Bacillus subtilis RNA polymerase (RNAP) elongation and HelD complexes, enabling analysis of the conformational changes that occur in RNAP driven by HelD interaction. HelD has a 2-armed structure which penetrates deep into the primary and secondary channels of RNA polymerase. One arm removes nucleic acids from the active site, and the other induces a large conformational change in the primary channel leading to removal and recycling of the stalled polymerase, representing a novel mechanism for recycling transcription complexes in bacteria. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21921.map.gz emd_21921.map.gz | 19.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21921-v30.xml emd-21921-v30.xml emd-21921.xml emd-21921.xml | 23 KB 23 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21921_fsc.xml emd_21921_fsc.xml | 10.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_21921.png emd_21921.png | 141 KB | ||
| Filedesc metadata |  emd-21921.cif.gz emd-21921.cif.gz | 8.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21921 http://ftp.pdbj.org/pub/emdb/structures/EMD-21921 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21921 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21921 | HTTPS FTP |
-Related structure data
| Related structure data |  6wvkMC  6wvjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-11052 (Title: Cryo-EM structure of Bacillus subtilis RNA Polymerase in complex with HelD EMPIAR-11052 (Title: Cryo-EM structure of Bacillus subtilis RNA Polymerase in complex with HelDData size: 2.8 TB Data #1: Unaligned multi-frame micrographs for Bacillus subtilis RNA Polymerase in complex with HelD [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21921.map.gz / Format: CCP4 / Size: 20.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21921.map.gz / Format: CCP4 / Size: 20.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | RNA Polymerase in complex with HelD | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : DNA-directed RNA polymerase complex with helD
| Entire | Name: DNA-directed RNA polymerase complex with helD |
|---|---|
| Components |
|
-Supramolecule #1: DNA-directed RNA polymerase complex with helD
| Supramolecule | Name: DNA-directed RNA polymerase complex with helD / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 442.06 KDa |
-Macromolecule #1: DNA-directed RNA polymerase subunit alpha
| Macromolecule | Name: DNA-directed RNA polymerase subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 34.842387 KDa |
| Sequence | String: MIEIEKPKIE TVEISDDAKF GKFVVEPLER GYGTTLGNSL RRILLSSLPG AAVTSIQIDG VLHEFSTIEG VVEDVTTIIL HIKKLALKI YSDEEKTLEI DVQGEGTVTA ADITHDSDVE ILNPDLHIAT LGENASFRVR LTAQRGRGYT PADANKRDDQ P IGVIPIDS ...String: MIEIEKPKIE TVEISDDAKF GKFVVEPLER GYGTTLGNSL RRILLSSLPG AAVTSIQIDG VLHEFSTIEG VVEDVTTIIL HIKKLALKI YSDEEKTLEI DVQGEGTVTA ADITHDSDVE ILNPDLHIAT LGENASFRVR LTAQRGRGYT PADANKRDDQ P IGVIPIDS IYTPVSRVSY QVENTRVGQV ANYDKLTLDV WTDGSTGPKE AIALGSKILT EHLNIFVGLT DEAQHAEIMV EK EEDQKEK VLEMTIEELD LSVRSYNCLK RAGINTVQEL ANKTEEDMMK VRNLGRKSLE EVKAKLEELG LGLRKDD UniProtKB: DNA-directed RNA polymerase subunit alpha |
-Macromolecule #2: DNA-directed RNA polymerase subunit beta
| Macromolecule | Name: DNA-directed RNA polymerase subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 133.847938 KDa |
| Sequence | String: MTGQLVQYGR HRQRRSYARI SEVLELPNLI EIQTSSYQWF LDEGLREMFQ DISPIEDFTG NLSLEFIDYS LGEPKYPVEE SKERDVTYS APLRVKVRLI NKETGEVKDQ DVFMGDFPIM TDTGTFIING AERVIVSQLV RSPSVYFSGK VDKNGKKGFT A TVIPNRGA ...String: MTGQLVQYGR HRQRRSYARI SEVLELPNLI EIQTSSYQWF LDEGLREMFQ DISPIEDFTG NLSLEFIDYS LGEPKYPVEE SKERDVTYS APLRVKVRLI NKETGEVKDQ DVFMGDFPIM TDTGTFIING AERVIVSQLV RSPSVYFSGK VDKNGKKGFT A TVIPNRGA WLEYETDAKD VVYVRIDRTR KLPVTVLLRA LGFGSDQEIL DLIGENEYLR NTLDKDNTEN SDKALLEIYE RL RPGEPPT VENAKSLLDS RFFDPKRYDL ANVGRYKINK KLHIKNRLFN QRLAETLVDP ETGEILAEKG QILDRRTLDK VLP YLENGI GFRKLYPNGG VVEDEVTLQS IKIFAPTDQE GEQVINVIGN AYIEEEIKNI TPADIISSIS YFFNLLHGVG DTDD IDHLG NRRLRSVGEL LQNQFRIGLS RMERVVRERM SIQDTNTITP QQLINIRPVI ASIKEFFGSS QLSQFMDQTN PLAEL THKR RLSALGPGGL TRERAGMEVR DVHYSHYGRM CPIETPEGPN IGLINSLSSY AKVNRFGFIE TPYRRVDPET GKVTGR IDY LTADEEDNYV VAQANARLDD EGAFIDDSIV ARFRGENTVV SRNRVDYMDV SPKQVVSAAT ACIPFLENDD SNRALMG AN MQRQAVPLMQ PEAPFVGTGM EYVSGKDSGA AVICKHPGIV ERVEAKNVWV RRYEEVDGQK VKGNLDKYSL LKFVRSNQ G TCYNQRPIVS VGDEVVKGEI LADGPSMELG ELALGRNVMV GFMTWDGYNY EDAIIMSERL VKDDVYTSIH IEEYESEAR DTKLGPEEIT RDIPNVGEDA LRNLDDRGII RIGAEVKDGD LLVGKVTPKG VTELTAEERL LHAIFGEKAR EVRDTSLRVP HGGGGIIHD VKVFNREDGD ELPPGVNQLV RVYIVQKRKI SEGDKMAGRH GNKGVISKIL PEEDMPYLPD GTPIDIMLNP L GVPSRMNI GQVLELHMGM AARYLGIHIA SPVFDGAREE DVWETLEEAG MSRDAKTVLY DGRTGEPFDN RVSVGIMYMI KL AHMVDDK LHARSTGPYS LVTQQPLGGK AQFGGQRFGE MEVWALEAYG AAYTLQEILT VKSDDVVGRV KTYEAIVKGD NVP EPGVPE SFKVLIKELQ SLGMDVKILS GDEEEIEMRD LEDEEDAKQA DGLALSGDEE PEETASADVE RDVVTKE UniProtKB: DNA-directed RNA polymerase subunit beta |
-Macromolecule #3: DNA-directed RNA polymerase subunit beta'
| Macromolecule | Name: DNA-directed RNA polymerase subunit beta' / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 134.444953 KDa |
| Sequence | String: MLDVNNFEYM NIGLASPDKI RSWSFGEVKK PETINYRTLK PEKDGLFCER IFGPTKDWEC HCGKYKRVRY KGVVCDRCGV EVTRAKVRR ERMGHIELAA PVSHIWYFKG IPSRMGLVLD MSPRALEEVI YFASYVVTDP ANTPLEKKQL LSEKEYRAYL D KYGNKFQA ...String: MLDVNNFEYM NIGLASPDKI RSWSFGEVKK PETINYRTLK PEKDGLFCER IFGPTKDWEC HCGKYKRVRY KGVVCDRCGV EVTRAKVRR ERMGHIELAA PVSHIWYFKG IPSRMGLVLD MSPRALEEVI YFASYVVTDP ANTPLEKKQL LSEKEYRAYL D KYGNKFQA SMGAEAIHKL LQDIDLVKEV DMLKEELKTS QGQRRTRAIK RLEVLEAFRN SGNKPSWMIL DVLPVIPPEL RP MVQLDGG RFATSDLNDL YRRVINRNNR LKRLLDLGAP SIIVQNEKRM LQEAVDALID NGRRGRPVTG PGNRPLKSLS HML KGKQGR FRQNLLGKRV DYSGRSVIVV GPHLKMYQCG LPKEMALELF KPFVMKELVE KGLAHNIKSA KRKIERVQPE VWDV LESVI KEHPVLLNRA PTLHRLGIQA FEPTLVEGRA IRLHPLVCTA YNADFDGDQM AVHVPLSAEA QAEARILMLA AQNIL NPKD GKPVVTPSQD MVLGNYYLTL ERAGAVGEGM VFKNTDEALL AYQNGYVHLH TRVAVAANSL KNVTFTEEQR SKLLIT TVG KLVFNEILPE SFPYMNEPTK SNIEEKTPDR FFLEKGADVK AVIAQQPINA PFKKGILGKI IAEIFKRFHI TETSKML DR MKNLGFKYST KAGITVGVSD IVVLDDKQEI LEEAQSKVDN VMKQFRRGLI TEEERYERVI SIWSAAKDVI QGKLMKSL D ELNPIYMMSD SGARGNASNF TQLAGMRGLM ANPAGRIIEL PIKSSFREGL TVLEYFISTH GARKGLADTA LKTADSGYL TRRLVDVAQD VIIRETDCGT DRGILAKPLK EGTETIERLE ERLIGRFARK QVKHPETGEV LVNENELIDE DKALEIVEAG IEEVWIRSA FTCNTPHGVC KRCYGRNLAT GSDVEVGEAV GIIAAQSIGE PGTQLTMRTF HTGGVAGDDI TQGLPRIQEL F EARNPKGQ ATITEIDGTV VEINEVRDKQ QEIVVQGAVE TRSYTAPYNS RLKVAEGDKI TRGQVLTEGS IDPKELLKVT DL TTVQEYL LHEVQKVYRM QGVEIGDKHV EVMVRQMLRK VRVIDAGDTD VLPGTLLDIH QFTEANKKVL LEGNRPATGR PVL LGITKA SLETDSFLSA ASFQETTRVL TDAAIKGKRD ELLGLKENVI IGKLVPAGTG MMKYRKVKPV SNVQPTDDMV PVE UniProtKB: DNA-directed RNA polymerase subunit beta' |
-Macromolecule #4: UPF0356 protein YkzG
| Macromolecule | Name: UPF0356 protein YkzG / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 8.263358 KDa |
| Sequence | String: MIYKVFYQEK ADEVPVREKT DSLYIEGVSE RDVRTKLKEK KFNIEFITPV DGAFLEYEQQ SENFKVLEL UniProtKB: DNA-directed RNA polymerase subunit epsilon |
-Macromolecule #5: DNA-directed RNA polymerase subunit omega
| Macromolecule | Name: DNA-directed RNA polymerase subunit omega / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.766921 KDa |
| Sequence | String: MLDPSIDSLM NKLDSKYTLV TVSARRAREM QIKKDQMIEH TISHKYVGKA LEEIDAGLLS FEKEDRE UniProtKB: DNA-directed RNA polymerase subunit omega |
-Macromolecule #6: DNA helicase IV
| Macromolecule | Name: DNA helicase IV / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 90.052516 KDa |
| Sequence | String: MNQQDKEWKE EQSRIDEVLK ELEKKERFLE TSAGGLKHDI IGLRKSFWED VKVNFDDAHE AIETMASIKQ QAELLSDREH NHRRMDQQL KRIHQLKKSP YFGRIDFIEN GEEQAERIYI GLASCLDEKE EHFLIYDWRA PISSLYYNYS PGKAEYEVPG E TIEGEMVL ...String: MNQQDKEWKE EQSRIDEVLK ELEKKERFLE TSAGGLKHDI IGLRKSFWED VKVNFDDAHE AIETMASIKQ QAELLSDREH NHRRMDQQL KRIHQLKKSP YFGRIDFIEN GEEQAERIYI GLASCLDEKE EHFLIYDWRA PISSLYYNYS PGKAEYEVPG E TIEGEMVL KRQFMIKNGT LKAMFNTDMT IGDEMLQEVL SHHSDTQMKN IVSTIQKEQN QIIRNEKSKI LIVQGAAGSG KT SAALQRV AYLLYRHRGV IDAGQIVLFS PNFLFNSYVS SVLPELGEEN MEQATFQEYI EHRLGRKFKC ESPFDQLEYC LTE TKGGDF PTRLAGITWK AGLSFQQFIN EYVTRLSSEG MIFKNIIFRG QKLITKEQIQ SYFYSLDQNH SIPNRMEQTA KWLL SELNK LEKKERRKDW VVHEAELLDK EDYLDVYKKL QERKRFSEST FNDYQREQQL LAAIIVKKAF KPLKQAVRLL AFLDV TQLY LQLFSGWGGK FQHEKMDAIG ELTRSAFTDN KLLYEDAAPF LYMQDLIEGR KKNTKIKHLF IDEAQDYSPF QMAYMR SIF PAASMTVLGD INQSIYAHTI NGDQRMDACF EDEPAEYVRL KRTYRSTRQI VEFTKAMLQD GADIEPFNRS GEMPLVV KT EGHESLCQKL AQEIGRLKKK GHETIAVICK TAHQCIQAHA HMSEYTDVRL IHKENQPFQK GVCVIPVYLA KGIEFDAV L VYDASEEHYH TEHDRRLLYT ACTRAMHMLA VFYTGEASPF VTAVPPHLYQ IAE UniProtKB: DNA helicase IV |
-Macromolecule #7: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 7 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #8: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 8 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.57 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.8 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Support film - topology: HOLEY / Support film - Film thickness: 50 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 32 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.02 kPa | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV Details: Sample loading volume ranged between 2 and 3 microlitres. Samples were blotted for 5 seconds prior to vitrification.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-60 / Number grids imaged: 1 / Number real images: 4331 / Average exposure time: 9.0 sec. / Average electron dose: 63.6 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 59500 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 0.6 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-6wvk: |
 Movie
Movie Controller
Controller
















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)