+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-21443 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | BurrH bound to DNA Origami Goniometer | ||||||||||||
 Map data Map data | Final refinement map for BurrH bound to DNA Goniometer tilt DNA | ||||||||||||
 Sample Sample |
| ||||||||||||
| Biological species |  Paraburkholderia rhizoxinica (bacteria) Paraburkholderia rhizoxinica (bacteria) | ||||||||||||
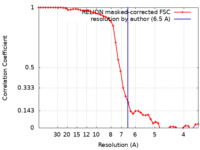
| Method | single particle reconstruction / cryo EM / Resolution: 6.5 Å | ||||||||||||
 Authors Authors | Aksel T / Yu Z / Cheng Y / Douglas SM | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Biotechnol / Year: 2021 Journal: Nat Biotechnol / Year: 2021Title: Molecular goniometers for single-particle cryo-electron microscopy of DNA-binding proteins. Authors: Tural Aksel / Zanlin Yu / Yifan Cheng / Shawn M Douglas /  Abstract: Correct reconstruction of macromolecular structure by cryo-electron microscopy (cryo-EM) relies on accurate determination of the orientation of single-particle images. For small (<100 kDa) DNA-binding proteins, obtaining particle images with sufficiently asymmetric features to correctly guide alignment is challenging. We apply DNA origami to construct molecular goniometers-instruments that precisely orient objects-and use them to dock a DNA-binding protein on a double-helix stage that has user-programmable tilt and rotation angles. We construct goniometers with 14 different stage configurations to orient and visualize the protein just above the cryo-EM grid surface. Each goniometer has a distinct barcode pattern that we use during particle classification to assign angle priors to the bound protein. We use goniometers to obtain a 6.5-Å structure of BurrH, an 82-kDa DNA-binding protein whose helical pseudosymmetry prevents accurate image orientation using traditional cryo-EM. Our approach should be adaptable to other DNA-binding proteins as well as small proteins fused to DNA-binding domains. #1:  Journal: Biorxiv / Year: 2020 Journal: Biorxiv / Year: 2020Title: Molecular goniometers for single-particle cryo-EM of DNA-binding proteins Authors: Aksel T / Yu Z / Cheng Y / Douglas SM | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_21443.map.gz emd_21443.map.gz | 6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-21443-v30.xml emd-21443-v30.xml emd-21443.xml emd-21443.xml | 18 KB 18 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_21443_fsc.xml emd_21443_fsc.xml | 4.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_21443.png emd_21443.png | 62.7 KB | ||
| Masks |  emd_21443_msk_1.map emd_21443_msk_1.map | 8 MB |  Mask map Mask map | |
| Others |  emd_21443_half_map_1.map.gz emd_21443_half_map_1.map.gz emd_21443_half_map_2.map.gz emd_21443_half_map_2.map.gz | 6 MB 6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-21443 http://ftp.pdbj.org/pub/emdb/structures/EMD-21443 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21443 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-21443 | HTTPS FTP |
-Validation report
| Summary document |  emd_21443_validation.pdf.gz emd_21443_validation.pdf.gz | 438 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_21443_full_validation.pdf.gz emd_21443_full_validation.pdf.gz | 437.6 KB | Display | |
| Data in XML |  emd_21443_validation.xml.gz emd_21443_validation.xml.gz | 10.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21443 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21443 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21443 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-21443 | HTTPS FTP |
-Related structure data
| Similar structure data | |
|---|---|
| EM raw data |  EMPIAR-10373 (Title: BurrH bound to DNA Origami Goniometer / Data size: 1.3 TB EMPIAR-10373 (Title: BurrH bound to DNA Origami Goniometer / Data size: 1.3 TBData #1: DNA Origami Goniometers in complex with BurrH DNA binding protein [micrographs - single frame] Data #2: BurrH particle set used for the final refinement [picked particles - multiframe - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_21443.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_21443.map.gz / Format: CCP4 / Size: 8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final refinement map for BurrH bound to DNA Goniometer tilt DNA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.82 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_21443_msk_1.map emd_21443_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: The first half map for BurrH bound to DNA Goniometer tilt DNA
| File | emd_21443_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The first half map for BurrH bound to DNA Goniometer tilt DNA | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: The second half map for BurrH bound to DNA Goniometer tilt DNA
| File | emd_21443_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The second half map for BurrH bound to DNA Goniometer tilt DNA | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : BurrH bound to DNA
| Entire | Name: BurrH bound to DNA |
|---|---|
| Components |
|
-Supramolecule #1: BurrH bound to DNA
| Supramolecule | Name: BurrH bound to DNA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Paraburkholderia rhizoxinica (bacteria) Paraburkholderia rhizoxinica (bacteria) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 81 KDa |
-Macromolecule #1: BurrH
| Macromolecule | Name: BurrH / type: protein_or_peptide / ID: 1 / Enantiomer: DEXTRO |
|---|---|
| Sequence | String: MGSTAFVDQD KQMANRLNLS PLERSKIEKQ YGGATTLAFI SNKQNELAQI LSRADILKIA SYDCAAHALQ AVLDCGPMLG KRGFSQSDI VKIAGNIGGA QALQAVLDLE SMLGKRGFSR DDIAKMAGNI GGAQTLQAVL DLESAFRERG FSQADIVKIA G NNGGAQAL ...String: MGSTAFVDQD KQMANRLNLS PLERSKIEKQ YGGATTLAFI SNKQNELAQI LSRADILKIA SYDCAAHALQ AVLDCGPMLG KRGFSQSDI VKIAGNIGGA QALQAVLDLE SMLGKRGFSR DDIAKMAGNI GGAQTLQAVL DLESAFRERG FSQADIVKIA G NNGGAQAL YSVLDVEPTL GKRGFSRADI VKIAGNTGGA QALHTVLDLE PALGKRGFSR IDIVKIAANN GGAQALHAVL DL GPTLREC GFSQATIAKI AGNIGGAQAL QMVLDLGPAL GKRGFSQATI AKIAGNIGGA QALQTVLDLE PALCERGFSQ ATI AKMAGN NGGAQALQTV LDLEPALRKR DFRQADIIKI AGNDGGAQAL QAVIEHGPTL RQHGFNLADI VKMAGNIGGA QALQ AVLDL KPVLDEHGFS QPDIVKMAGN IGGAQALQAV LSLGPALRER GFSQPDIVKI AGNTGGAQAL QAVLDLELTL VEHGF SQPD IVRITGNRGG AQALQAVLAL ELTLRERGFS QPDIVKIAGN SGGAQALQAV LDLELTFRER GFSQADIVKI AGNDGG TQA LHAVLDLERM LGERGFSRAD IVNVAGNNGG AQALKAVLEH EATLNERGFS RADIVKIAGN GGGAQALKAV LEHEATL DE RGFSRADIVR IAGNGGGAQA LKAVLEHGPT LNERGFNLTD IVEMAANSGG AQALKAVLEH GPTLRQRGLS LIDIVEIA S NGGAQALKAV LKYGPVLMQA GRSNEEIVHV AARRGGAGRI RKMVAPLLER QWGSGHHHHH H |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
| Details | DNA Origami goniometer complexed with BurrH. Total concentration comprises the concentrations of both. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 56.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 20.0 µm / Nominal defocus min: 5.0 µm / Nominal magnification: 22000 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: RMSD |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)