[English] 日本語
 Yorodumi
Yorodumi- EMDB-20798: Integrin alpha-v beta-8 in complex with latent TGF-beta, Conforma... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-20798 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
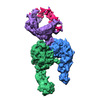
| Title | Integrin alpha-v beta-8 in complex with latent TGF-beta, Conformation ii (Primary map: sharpened. Additional map: unsharpened.) | ||||||||||||||||||||||||
 Map data Map data | Integrin alpha-v beta-8 in complex with latent TGF-beta, Conformation ii, sharpened map | ||||||||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.6 Å | ||||||||||||||||||||||||
 Authors Authors | Campbell MG / Cormier A / Cheng Y / Nishimura SL | ||||||||||||||||||||||||
| Funding support |  United States, 7 items United States, 7 items
| ||||||||||||||||||||||||
 Citation Citation |  Journal: Cell / Year: 2020 Journal: Cell / Year: 2020Title: Cryo-EM Reveals Integrin-Mediated TGF-β Activation without Release from Latent TGF-β. Authors: Melody G Campbell / Anthony Cormier / Saburo Ito / Robert I Seed / Andrew J Bondesson / Jianlong Lou / James D Marks / Jody L Baron / Yifan Cheng / Stephen L Nishimura /  Abstract: Integrin αvβ8 binds with exquisite specificity to latent transforming growth factor-β (L-TGF-β). This binding is essential for activating L-TGF-β presented by a variety of cell types. ...Integrin αvβ8 binds with exquisite specificity to latent transforming growth factor-β (L-TGF-β). This binding is essential for activating L-TGF-β presented by a variety of cell types. Inhibiting αvβ8-mediated TGF-β activation blocks immunosuppressive regulatory T cell differentiation, which is a potential therapeutic strategy in cancer. Using cryo-electron microscopy, structure-guided mutagenesis, and cell-based assays, we reveal the binding interactions between the entire αvβ8 ectodomain and its intact natural ligand, L-TGF-β, as well as two different inhibitory antibody fragments to understand the structural underpinnings of αvβ8 binding specificity and TGF-β activation. Our studies reveal a mechanism of TGF-β activation where mature TGF-β signals within the confines of L-TGF-β and the release and diffusion of TGF-β are not required. The structural details of this mechanism provide a rational basis for therapeutic strategies to inhibit αvβ8-mediated L-TGF-β activation. | ||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_20798.map.gz emd_20798.map.gz | 1.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-20798-v30.xml emd-20798-v30.xml emd-20798.xml emd-20798.xml | 14.6 KB 14.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_20798.png emd_20798.png | 93.1 KB | ||
| Others |  emd_20798_additional.map.gz emd_20798_additional.map.gz emd_20798_additional_1.map.gz emd_20798_additional_1.map.gz | 52 MB 52 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-20798 http://ftp.pdbj.org/pub/emdb/structures/EMD-20798 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20798 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-20798 | HTTPS FTP |
-Related structure data
| Related structure data |  6ujaC  6ujbC  6ujcC C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10343 (Title: CryoEM dataset containing multiple conformations of the asymmetric αVβ8 integrin bound to latent TGF-β on a holey carbon grid (strongly preferred orientations) EMPIAR-10343 (Title: CryoEM dataset containing multiple conformations of the asymmetric αVβ8 integrin bound to latent TGF-β on a holey carbon grid (strongly preferred orientations)Data size: 1.2 TB Data #1: Unaligned 80-frame movies of αVβ8 integrin bound to latent TGF-β on a holey carbon grid [micrographs - multiframe] Data #2: Dose-weighted aligned micrographs of αVβ8 integrin bound to latent TGF-β on a holey carbon grid [micrographs - single frame] Data #3: Dose-weighted aligned particle stacks of αVβ8 integrin bound to latent TGF-β on a holey carbon grid [picked particles - single frame - processed])  EMPIAR-10344 (Title: CryoEM dataset containing multiple conformations of the asymmetric αVβ8 integrin bound to latent TGF-β on a graphene oxide grid (preferred orientations) EMPIAR-10344 (Title: CryoEM dataset containing multiple conformations of the asymmetric αVβ8 integrin bound to latent TGF-β on a graphene oxide grid (preferred orientations)Data size: 2.2 TB Data #1: Unaligned 80-frame movies of αVβ8 integrin bound to latent TGF-β on a graphene oxide grid [micrographs - multiframe] Data #2: Dose-weighted aligned micrographs of αVβ8 integrin bound to latent TGF-β on a graphene oxide grid [micrographs - single frame] Data #3: Dose-weighted aligned particle stacks of αVβ8 integrin bound to latent TGF-β on a graphene oxide grid [picked particles - single frame - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_20798.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_20798.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Integrin alpha-v beta-8 in complex with latent TGF-beta, Conformation ii, sharpened map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.345 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
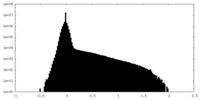
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: Integrin alpha-v beta-8 in complex with latent TGF-beta,...
| File | emd_20798_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Integrin alpha-v beta-8 in complex with latent TGF-beta, Conformation ii (Additional unsharpened map.) | ||||||||||||
| Projections & Slices |
| ||||||||||||
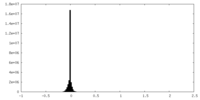
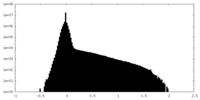
| Density Histograms |
-Additional map: Integrin alpha-v beta-8 in complex with latent TGF-beta,...
| File | emd_20798_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Integrin alpha-v beta-8 in complex with latent TGF-beta, Conformation ii (Additional unsharpened map.) | ||||||||||||
| Projections & Slices |
| ||||||||||||
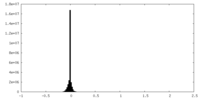
| Density Histograms |
- Sample components
Sample components
-Entire : Binary complex of Integrin alpha-v beta-8 with Latent TGF-beta-1 ...
| Entire | Name: Binary complex of Integrin alpha-v beta-8 with Latent TGF-beta-1 (L-TGF-b1) |
|---|---|
| Components |
|
-Supramolecule #1: Binary complex of Integrin alpha-v beta-8 with Latent TGF-beta-1 ...
| Supramolecule | Name: Binary complex of Integrin alpha-v beta-8 with Latent TGF-beta-1 (L-TGF-b1) type: complex / ID: 1 / Parent: 0 |
|---|---|
| Molecular weight | Theoretical: 260 KDa |
-Supramolecule #2: alpha-v beta-8 integrin
| Supramolecule | Name: alpha-v beta-8 integrin / type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
-Supramolecule #3: Transforming Growth Factor Beta-1 proprotein
| Supramolecule | Name: Transforming Growth Factor Beta-1 proprotein / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: HEK293A Homo sapiens (human) / Recombinant cell: HEK293A |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Details: unspecified |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 3.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 32689 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)