[English] 日本語
 Yorodumi
Yorodumi- EMDB-17813: Cryo-EM structure of human DNA polymerase alpha-primase at initia... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human DNA polymerase alpha-primase at initiation II | |||||||||
 Map data Map data | sharpened final map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | DNA polymerase / complex / DNA BINDING PROTEIN | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.03 Å | |||||||||
 Authors Authors | Yin Z / Pellegrini L | |||||||||
| Funding support |  United Kingdom, 1 items United Kingdom, 1 items
| |||||||||
 Citation Citation |  Journal: FEBS J / Year: 2024 Journal: FEBS J / Year: 2024Title: CryoEM insights into RNA primer synthesis by the human primosome. Authors: Zhan Yin / Mairi L Kilkenny / De-Sheng Ker / Luca Pellegrini /  Abstract: Eukaryotic DNA replication depends on the primosome - a complex of DNA polymerase alpha (Pol α) and primase - to initiate DNA synthesis by polymerisation of an RNA-DNA primer. Primer synthesis ...Eukaryotic DNA replication depends on the primosome - a complex of DNA polymerase alpha (Pol α) and primase - to initiate DNA synthesis by polymerisation of an RNA-DNA primer. Primer synthesis requires the tight coordination of primase and polymerase activities. Recent cryo-electron microscopy (cryoEM) analyses have elucidated the extensive conformational transitions required for RNA primer handover between primase and Pol α and primer elongation by Pol α. Because of the intrinsic flexibility of the primosome, however, structural information about the initiation of RNA primer synthesis is still lacking. Here, we capture cryoEM snapshots of the priming reaction to reveal the conformational trajectory of the human primosome that brings DNA primase subunits 1 and 2 (PRIM1 and PRIM2, respectively) together, poised for RNA synthesis. Furthermore, we provide experimental evidence for the continuous association of primase subunit PRIM2 with the RNA primer during primer synthesis, and for how both initiation and termination of RNA primer polymerisation are licenced by specific rearrangements of DNA polymerase alpha catalytic subunit (POLA1), the polymerase subunit of Pol α. Our findings fill a critical gap in our understanding of the conformational changes that underpin the synthesis of the RNA primer by the primosome. Together with existing evidence, they provide a complete description of the structural dynamics of the human primosome during DNA replication initiation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_17813.map.gz emd_17813.map.gz | 197.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-17813-v30.xml emd-17813-v30.xml emd-17813.xml emd-17813.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_17813_fsc.xml emd_17813_fsc.xml | 12.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_17813.png emd_17813.png | 50.7 KB | ||
| Masks |  emd_17813_msk_1.map emd_17813_msk_1.map | 209.3 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-17813.cif.gz emd-17813.cif.gz | 3.8 KB | ||
| Others |  emd_17813_additional_1.map.gz emd_17813_additional_1.map.gz emd_17813_half_map_1.map.gz emd_17813_half_map_1.map.gz emd_17813_half_map_2.map.gz emd_17813_half_map_2.map.gz | 104.6 MB 194.5 MB 194.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-17813 http://ftp.pdbj.org/pub/emdb/structures/EMD-17813 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17813 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-17813 | HTTPS FTP |
-Validation report
| Summary document |  emd_17813_validation.pdf.gz emd_17813_validation.pdf.gz | 940.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_17813_full_validation.pdf.gz emd_17813_full_validation.pdf.gz | 939.9 KB | Display | |
| Data in XML |  emd_17813_validation.xml.gz emd_17813_validation.xml.gz | 21.4 KB | Display | |
| Data in CIF |  emd_17813_validation.cif.gz emd_17813_validation.cif.gz | 27.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17813 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17813 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17813 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-17813 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_17813.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_17813.map.gz / Format: CCP4 / Size: 209.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened final map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.066 Å | ||||||||||||||||||||||||||||||||||||
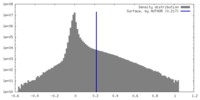
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_17813_msk_1.map emd_17813_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
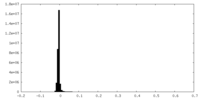
| Density Histograms |
-Additional map: non-sharpened final map
| File | emd_17813_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | non-sharpened final map | ||||||||||||
| Projections & Slices |
| ||||||||||||
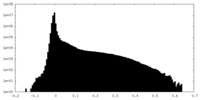
| Density Histograms |
-Half map: #1
| File | emd_17813_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
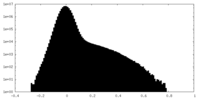
| Density Histograms |
-Half map: #2
| File | emd_17813_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : DNA polymerase alpha-primase
| Entire | Name: DNA polymerase alpha-primase |
|---|---|
| Components |
|
-Supramolecule #1: DNA polymerase alpha-primase
| Supramolecule | Name: DNA polymerase alpha-primase / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 46.05 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.6 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)