[English] 日本語
 Yorodumi
Yorodumi- EMDB-16041: CryoEM structure of quinol-dependent Nitric Oxide Reductase (qNOR... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
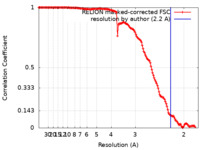
| Title | CryoEM structure of quinol-dependent Nitric Oxide Reductase (qNOR) from Alcaligenes xylosoxidans at 2.2 A resolution | |||||||||
 Map data Map data | main map used for model building | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | quinol-dependent Nitric Oxide Reductase / proton transfer / quinol binding / ubiquinol oxidase / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnitric oxide reductase (cytochrome c) / nitric oxide reductase activity / cytochrome-c oxidase activity / aerobic respiration / heme binding / membrane Similarity search - Function | |||||||||
| Biological species |  Achromobacter xylosoxidans (bacteria) Achromobacter xylosoxidans (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.2 Å | |||||||||
 Authors Authors | Flynn A / Antonyuk SV / Eady RR / Muench SP / Hasnain SS | |||||||||
| Funding support |  United Kingdom, 2 items United Kingdom, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: A 2.2 Å cryoEM structure of a quinol-dependent NO Reductase shows close similarity to respiratory oxidases. Authors: Alex J Flynn / Svetlana V Antonyuk / Robert R Eady / Stephen P Muench / S Samar Hasnain /  Abstract: Quinol-dependent nitric oxide reductases (qNORs) are considered members of the respiratory heme-copper oxidase superfamily, are unique to bacteria, and are commonly found in pathogenic bacteria where ...Quinol-dependent nitric oxide reductases (qNORs) are considered members of the respiratory heme-copper oxidase superfamily, are unique to bacteria, and are commonly found in pathogenic bacteria where they play a role in combating the host immune response. qNORs are also essential enzymes in the denitrification pathway, catalysing the reduction of nitric oxide to nitrous oxide. Here, we determine a 2.2 Å cryoEM structure of qNOR from Alcaligenes xylosoxidans, an opportunistic pathogen and a denitrifying bacterium of importance in the nitrogen cycle. This high-resolution structure provides insight into electron, substrate, and proton pathways, and provides evidence that the quinol binding site not only contains the conserved His and Asp residues but also possesses a critical Arg (Arg720) observed in cytochrome bo, a respiratory quinol oxidase. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16041.map.gz emd_16041.map.gz | 14.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16041-v30.xml emd-16041-v30.xml emd-16041.xml emd-16041.xml | 22 KB 22 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_16041_fsc.xml emd_16041_fsc.xml | 14 KB | Display |  FSC data file FSC data file |
| Images |  emd_16041.png emd_16041.png | 112.4 KB | ||
| Masks |  emd_16041_msk_1.map emd_16041_msk_1.map | 244.1 MB |  Mask map Mask map | |
| Others |  emd_16041_half_map_1.map.gz emd_16041_half_map_1.map.gz emd_16041_half_map_2.map.gz emd_16041_half_map_2.map.gz | 192.5 MB 192.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16041 http://ftp.pdbj.org/pub/emdb/structures/EMD-16041 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16041 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16041 | HTTPS FTP |
-Related structure data
| Related structure data |  8bgwMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16041.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16041.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | main map used for model building | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.91 Å | ||||||||||||||||||||||||||||||||||||
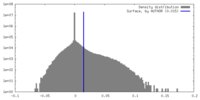
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16041_msk_1.map emd_16041_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
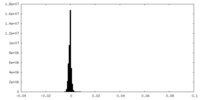
| Density Histograms |
-Half map: half map used in refinement
| File | emd_16041_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map used in refinement | ||||||||||||
| Projections & Slices |
| ||||||||||||
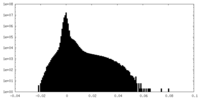
| Density Histograms |
-Half map: half map used for cross validation
| File | emd_16041_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map used for cross validation | ||||||||||||
| Projections & Slices |
| ||||||||||||
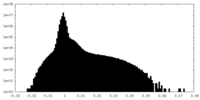
| Density Histograms |
- Sample components
Sample components
-Entire : quinol-dependent Nitric Oxide Reductase
| Entire | Name: quinol-dependent Nitric Oxide Reductase |
|---|---|
| Components |
|
-Supramolecule #1: quinol-dependent Nitric Oxide Reductase
| Supramolecule | Name: quinol-dependent Nitric Oxide Reductase / type: cell / ID: 1 / Parent: 0 / Macromolecule list: #1 / Details: quinol-dependent Nitric Oxide Reductase |
|---|---|
| Source (natural) | Organism:  Achromobacter xylosoxidans (bacteria) Achromobacter xylosoxidans (bacteria) |
-Macromolecule #1: Nitric oxide reductase subunit B
| Macromolecule | Name: Nitric oxide reductase subunit B / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: nitric oxide reductase (cytochrome c) |
|---|---|
| Source (natural) | Organism:  Achromobacter xylosoxidans (bacteria) Achromobacter xylosoxidans (bacteria) |
| Molecular weight | Theoretical: 84.724867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGPYRRLWFT LIAVLAVTFA LLGFYGGEVY RQAPPIPEEV ASADGTRLFG RDDILDGQTA WQSIGGMQLG SIWGHGAYQA PDWTADWLH RELMAWLDLA ARDAHGRDYG QLDAPAQAAL REQLKAEYRA NRADAAGGKL TLSPRRAQAV AQTEAYYDQL F SDAPALHR ...String: MGPYRRLWFT LIAVLAVTFA LLGFYGGEVY RQAPPIPEEV ASADGTRLFG RDDILDGQTA WQSIGGMQLG SIWGHGAYQA PDWTADWLH RELMAWLDLA ARDAHGRDYG QLDAPAQAAL REQLKAEYRA NRADAAGGKL TLSPRRAQAV AQTEAYYDQL F SDAPALHR SRENYAMKEN TLPDANRRRQ MTHFFFWTAW AAATEREGTS VTYTNNWPHE PLIGNHPSSE NVMWSIISVV VL LAGIGLL IWAWAFLRGK EEDEPPAPAR DPLTTFALTP SQRALGKYLF LVVALFGFQV LLGGFTAHYT VEGQKFYGID LSQ WFPYSL VRTWHIQSAL FWIATGFLAA GLFLAPLING GRDPKYQKAG VDILFWALVL VVVGSFAGNY LAIAQIMPPD LNFW LGHQG YEYVDLGRLW QIGKFAGICF WLVLMLRGIV PALRTPGGDK NLLALLTASV GAIGLFYGAG FFYGERTHLT VMEYW RWWI VHLWVEGFFE VFATTALAFI FSTLGLVSRR MATTASLASA SLFMLGGIPG TFHHLYFAGT TTPVMAVGAS FSALEV VPL IVLGHEAWEN WRLKTRAPWM ENLKWPLMCF VAVAFWNMLG AGVFGFMINP PVSLYYIQGL NTTPVHAHAA LFGVYGF LA LGFTLLVLRY IRPQYALSPG LMKLAFWGLN LGLALMIFTS LLPIGLIQFH ASVSEGMWYA RSEAFMQQDI LKTLRWGR T FGDVVFLLGA LAMVVQVILG LLSGKPAAAE PVLRAEPARR UniProtKB: Nitric oxide reductase subunit B |
-Macromolecule #2: PROTOPORPHYRIN IX CONTAINING FE
| Macromolecule | Name: PROTOPORPHYRIN IX CONTAINING FE / type: ligand / ID: 2 / Number of copies: 4 / Formula: HEM |
|---|---|
| Molecular weight | Theoretical: 616.487 Da |
| Chemical component information |  ChemComp-HEM: |
-Macromolecule #3: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Macromolecule #4: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #5: decyl 4-O-alpha-D-glucopyranosyl-1-thio-beta-D-glucopyranoside
| Macromolecule | Name: decyl 4-O-alpha-D-glucopyranosyl-1-thio-beta-D-glucopyranoside type: ligand / ID: 5 / Number of copies: 2 / Formula: 10M |
|---|---|
| Molecular weight | Theoretical: 498.628 Da |
| Chemical component information | 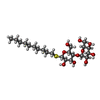 ChemComp-10M: |
-Macromolecule #6: UBIQUINONE-1
| Macromolecule | Name: UBIQUINONE-1 / type: ligand / ID: 6 / Number of copies: 2 / Formula: UQ1 |
|---|---|
| Molecular weight | Theoretical: 250.29 Da |
| Chemical component information | 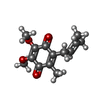 ChemComp-UQ1: |
-Macromolecule #7: (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANO...
| Macromolecule | Name: (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE type: ligand / ID: 7 / Number of copies: 13 / Formula: LOP |
|---|---|
| Molecular weight | Theoretical: 661.89 Da |
| Chemical component information |  ChemComp-LOP: |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 449 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 7 / Component - Concentration: 50.0 mM / Component - Formula: Tris-HCL / Component - Name: Tris-HCL / Details: 50mM Tris-HCl, 150mM NaCl, 0.05% DTM |
| Grid | Model: Quantifoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Sample at 3 mg/mL was applied to glow-discharged Quantifoil Au R1.2/1.3 holey carbo grids. |
| Details | Sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 80.0 K / Max: 114.0 K |
| Specialist optics | Energy filter - Name: TFS Selectris / Energy filter - Slit width: 10 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 1 / Number real images: 5466 / Average exposure time: 6.11 sec. / Average electron dose: 34.9 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.7 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: RECIPROCAL / Protocol: RIGID BODY FIT / Overall B value: 47 / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model |  PDB-8bgw: |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)