+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | COPII inner coat reprocessed with relion4.0 | |||||||||
 Map data Map data | COPII inner coat | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | COPII / Sec23 Sec24 Sar1 / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationAntigen Presentation: Folding, assembly and peptide loading of class I MHC / Cargo concentration in the ER / regulation of COPII vesicle coating / positive regulation of ER to Golgi vesicle-mediated transport / nuclear envelope organization / COPII-coated vesicle budding / COPII-mediated vesicle transport / mitochondria-associated endoplasmic reticulum membrane contact site / positive regulation of protein exit from endoplasmic reticulum / vesicle organization ...Antigen Presentation: Folding, assembly and peptide loading of class I MHC / Cargo concentration in the ER / regulation of COPII vesicle coating / positive regulation of ER to Golgi vesicle-mediated transport / nuclear envelope organization / COPII-coated vesicle budding / COPII-mediated vesicle transport / mitochondria-associated endoplasmic reticulum membrane contact site / positive regulation of protein exit from endoplasmic reticulum / vesicle organization / COPII vesicle coat / membrane organization / mitochondrial fission / mitochondrial membrane organization / endoplasmic reticulum exit site / endoplasmic reticulum to Golgi vesicle-mediated transport / intracellular protein transport / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / Golgi membrane / GTPase activity / endoplasmic reticulum membrane / GTP binding / endoplasmic reticulum / Golgi apparatus / mitochondrion / zinc ion binding Similarity search - Function | |||||||||
| Biological species |   | |||||||||
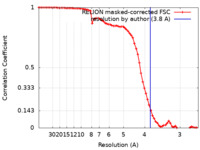
| Method | subtomogram averaging / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Zanetti G / Pyle E | |||||||||
| Funding support | European Union, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: A Bayesian approach to single-particle electron cryo-tomography in RELION-4.0. Authors: Jasenko Zivanov / Joaquín Otón / Zunlong Ke / Andriko von Kügelgen / Euan Pyle / Kun Qu / Dustin Morado / Daniel Castaño-Díez / Giulia Zanetti / Tanmay A M Bharat / John A G Briggs / Sjors H W Scheres /     Abstract: We present a new approach for macromolecular structure determination from multiple particles in electron cryo-tomography (cryo-ET) data sets. Whereas existing subtomogram averaging approaches are ...We present a new approach for macromolecular structure determination from multiple particles in electron cryo-tomography (cryo-ET) data sets. Whereas existing subtomogram averaging approaches are based on 3D data models, we propose to optimise a regularised likelihood target that approximates a function of the 2D experimental images. In addition, analogous to Bayesian polishing and contrast transfer function (CTF) refinement in single-particle analysis, we describe the approaches that exploit the increased signal-to-noise ratio in the averaged structure to optimise tilt-series alignments, beam-induced motions of the particles throughout the tilt-series acquisition, defoci of the individual particles, as well as higher-order optical aberrations of the microscope. Implementation of our approaches in the open-source software package RELION aims to facilitate their general use, particularly for those researchers who are already familiar with its single-particle analysis tools. We illustrate for three applications that our approaches allow structure determination from cryo-ET data to resolutions sufficient for de novo atomic modelling. #1:  Journal: Biorxiv / Year: 2022 Journal: Biorxiv / Year: 2022Title: A Bayesian approach to single-particle electron cryo-tomography in RELION-4.0 Authors: Zivanov J / Oton J / Ke Z / von Kugelgen A / Pyle E / Qu K / Morado D / Castano-Diez D / Zanetti G / Bharat TAM / Briggs JAG / Scheres SHW | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15949.map.gz emd_15949.map.gz | 18.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15949-v30.xml emd-15949-v30.xml emd-15949.xml emd-15949.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_15949_fsc.xml emd_15949_fsc.xml | 7 KB | Display |  FSC data file FSC data file |
| Images |  emd_15949.png emd_15949.png | 156 KB | ||
| Masks |  emd_15949_msk_1.map emd_15949_msk_1.map | 28.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-15949.cif.gz emd-15949.cif.gz | 6.5 KB | ||
| Others |  emd_15949_half_map_1.map.gz emd_15949_half_map_1.map.gz emd_15949_half_map_2.map.gz emd_15949_half_map_2.map.gz | 14.1 MB 14.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15949 http://ftp.pdbj.org/pub/emdb/structures/EMD-15949 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15949 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15949 | HTTPS FTP |
-Related structure data
| Related structure data |  8bshMC  8bqeC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15949.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15949.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | COPII inner coat | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_15949_msk_1.map emd_15949_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half2
| File | emd_15949_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half1
| File | emd_15949_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : COPII coat assembled on membrane, inner coat
| Entire | Name: COPII coat assembled on membrane, inner coat |
|---|---|
| Components |
|
-Supramolecule #1: COPII coat assembled on membrane, inner coat
| Supramolecule | Name: COPII coat assembled on membrane, inner coat / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|
-Supramolecule #2: Protein transport protein SEC23
| Supramolecule | Name: Protein transport protein SEC23 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1, #4 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Protein transport protein SEC24
| Supramolecule | Name: Protein transport protein SEC24 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #4: Small COPII coat GTPase SAR1
| Supramolecule | Name: Small COPII coat GTPase SAR1 / type: complex / ID: 4 / Parent: 1 / Macromolecule list: #3, #5 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: inner COPII coat complex
| Macromolecule | Name: inner COPII coat complex / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Spodoptera (butterflies/moths) Spodoptera (butterflies/moths) |
| Sequence | String: DFETNEDING VRFTWNVFPS TRSDANSNVV PVGCLYTPLK EYDELNVAPY NPVVCSGPHC KSILNPYCVI DPRNSSWSCP ICNSRNHLPP QYTNLSQENM PLELQSTTIE YITNKPVTVP PIFFFVVDLT SETENLDSLK ESIITSLSLL PPNALIGLIT YGNVVQLHDL ...String: DFETNEDING VRFTWNVFPS TRSDANSNVV PVGCLYTPLK EYDELNVAPY NPVVCSGPHC KSILNPYCVI DPRNSSWSCP ICNSRNHLPP QYTNLSQENM PLELQSTTIE YITNKPVTVP PIFFFVVDLT SETENLDSLK ESIITSLSLL PPNALIGLIT YGNVVQLHDL SSETIDRCNV FRGDREYQLE ALTEMLTGQK PTGPGGAASH LPNAMNKVTP FSLNRFFLP LEQVEFKLNQ LLENLSPDQW SVPAGHRPLR ATGSALNIAS LLLQGCYKNI PARIILFASG PGTVAPGLIV NSELKDPLRS HHDIDSDHAQ HYKKACKFYN QIAQRVAANG HTVDIFAGCY DQIGMSEMKQ LTDSTGGVLL LTDAFSTAIF KQSYLRLFAK DEEGYLKMAF NGNMAVKTSK DLKVQGLIGH ASAVKKTDAN NISESEIGIG A TSTWKMAS LSPYHSYAIF FEIANTAANS NPMMSAPGSA DRPHLAYTQF ITTYQHSSGT NRIRVTTVAN QLLPFGTPAI AASFDQEAAA VLMARIAVHK AETDDGADVI RWLDRTLIKL CQKYADYNKD DPQSFRLAPN FSLYPQFTYY LRRSQFLSVF NNSPDETAFY RHIFTREDTT NSLIMIQPTL TSFSMEDDPQ PVLLDSISVK PNTILLLDTF FF ILIYHGE QIAQWRKAGY QDDPQYADFK ALLEEPKLEA AELLVDRFPL PRFIDTEAGG SQARFLLSKL NPSDNYQDMA RGGSTIVLTD DVSLQNFMTH LQQVAVSGQA MSHHKKRVY PQAQLQYGQN ATPLQQPAQF MPPQDPAAAG MSYGQMGMPP QGAVPSMGQQ QFLTPAQEQL HQQIDQATTS MNDMHLHNVP LVDPNAYMQP QVPVQMGTPL QQQQQPMAAP AYGQPSAAMG QNMRPMNQLY PIDLLTELPP PITDLTLPPP PLVIPPERML VPSELSNASP DYIRSTLNAV PKNSSLLKKS KLPFGLVIRP YQHLYDDIDP P PLNEDGLI VRCRRCRSYM NPFVTFIEQG RRWRCNFCRL ANDVPMQMDQ SDPNDPKSRY DRNEIKCAVM EYMAPKEYTL RQPPPATYCF LIDVSQSSIK SGLLATTINT LLQNLDSIPN HDERTRISIL CVDNAIHYFK IPLDSENNEE SADQINMMDI ADLEEPFLPR PNSMVVSLKA CRQNIETLLT KIPQIFQSNL ITNFALGPAL KSAYHLIGGV GG KIIVVSG TLPNLGIGKL QRRNESGVVN TSKETAQLLS CQDSFYKNFT IDCSKVQITV DLFLASEDYM DVASLSNLSR FTAGQTHFYP GFSGKNPNDI VKFSTEFAKH ISMDFCMETV MRARGSTGLR MSRFYGHFFN RSSDLCAFST MPRDQSYLFE VNVDESIMAD YCYVQVAVLL SLNNSQRRIR IITLAMPTTE SLAEVYASAD QLAIASFYNS KAV EKALNS SLDDARVLIN KSVQDILATY KKEIVVSNTA GGAPLRLCAN LRMFPLLMHS LTKHMAFRSG IVPSDHRASA LNNLESLPLK YLIKNIYPDV YSLHDMADEA GLPVQTEDGE ATGTIVLPQP INATSSLFER YGLYLIDNGN ELFLWMGGDA VPALVFDVFG TQDIFDIPIG KQEIPVVENS EFNQRVRNII NQLRNHDDVI TYQSLYIVRG ASLS EPVNH ASAREVATLR LWASSTLVED KILNNESYRE FLQIMKARIS K MAGWDIFG WFRDVLASLG LWNKHGKLLF LGLDNAGKTT LLHMLKNDRL ATLQPTWHPT SEELAIGNIK FTTFDLGGHI QARRLWKDYF PEVNGIVFLV DAADPERFDE ARVELDALFN IAELKDVPFV ILGNKIDAPN AVSEAELRSA LGLLNTTGSQ RIEGQRPVEV FMCSVVMRNG YLEAFQWLSQ YI |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Buffer | pH: 6.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 3.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 3.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)