[English] 日本語
 Yorodumi
Yorodumi- EMDB-16209: HIV-1 CA-SP1 subtomogram average with Relion4 from EMPIAR-10164 d... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | HIV-1 CA-SP1 subtomogram average with Relion4 from EMPIAR-10164 dataset, 3.0 A from the full dataset | ||||||||||||||||||
 Map data Map data | HIV-1 CA-SP1 subtomogram average, from the full dataset, relion4.0 tomo | ||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | HIV-1 CA-SP1 / data from EMPIAR-10164 / VIRAL PROTEIN | ||||||||||||||||||
| Biological species |   Human immunodeficiency virus 1 Human immunodeficiency virus 1 | ||||||||||||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 3.0 Å | ||||||||||||||||||
 Authors Authors | Ke Z / Briggs JAG / Scheres SHW | ||||||||||||||||||
| Funding support |  United Kingdom, European Union, United Kingdom, European Union,  Germany, 5 items Germany, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2022 Journal: Elife / Year: 2022Title: A Bayesian approach to single-particle electron cryo-tomography in RELION-4.0. Authors: Jasenko Zivanov / Joaquín Otón / Zunlong Ke / Andriko von Kügelgen / Euan Pyle / Kun Qu / Dustin Morado / Daniel Castaño-Díez / Giulia Zanetti / Tanmay A M Bharat / John A G Briggs / Sjors H W Scheres /     Abstract: We present a new approach for macromolecular structure determination from multiple particles in electron cryo-tomography (cryo-ET) data sets. Whereas existing subtomogram averaging approaches are ...We present a new approach for macromolecular structure determination from multiple particles in electron cryo-tomography (cryo-ET) data sets. Whereas existing subtomogram averaging approaches are based on 3D data models, we propose to optimise a regularised likelihood target that approximates a function of the 2D experimental images. In addition, analogous to Bayesian polishing and contrast transfer function (CTF) refinement in single-particle analysis, we describe the approaches that exploit the increased signal-to-noise ratio in the averaged structure to optimise tilt-series alignments, beam-induced motions of the particles throughout the tilt-series acquisition, defoci of the individual particles, as well as higher-order optical aberrations of the microscope. Implementation of our approaches in the open-source software package RELION aims to facilitate their general use, particularly for those researchers who are already familiar with its single-particle analysis tools. We illustrate for three applications that our approaches allow structure determination from cryo-ET data to resolutions sufficient for de novo atomic modelling. | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_16209.map.gz emd_16209.map.gz | 25.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-16209-v30.xml emd-16209-v30.xml emd-16209.xml emd-16209.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
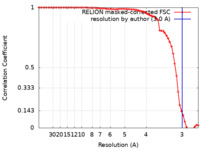
| FSC (resolution estimation) |  emd_16209_fsc.xml emd_16209_fsc.xml | 6.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_16209.png emd_16209.png | 182.6 KB | ||
| Masks |  emd_16209_msk_1.map emd_16209_msk_1.map | 27 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-16209.cif.gz emd-16209.cif.gz | 4.2 KB | ||
| Others |  emd_16209_half_map_1.map.gz emd_16209_half_map_1.map.gz emd_16209_half_map_2.map.gz emd_16209_half_map_2.map.gz | 19.7 MB 19.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-16209 http://ftp.pdbj.org/pub/emdb/structures/EMD-16209 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16209 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-16209 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_16209.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_16209.map.gz / Format: CCP4 / Size: 27 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HIV-1 CA-SP1 subtomogram average, from the full dataset, relion4.0 tomo | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_16209_msk_1.map emd_16209_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
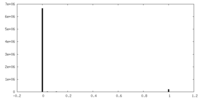
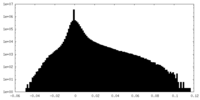
| Density Histograms |
-Half map: HIV-1 CA-SP1 subtomogram average, from the full dataset,...
| File | emd_16209_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HIV-1 CA-SP1 subtomogram average, from the full dataset, relion4.0 tomo, half1 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
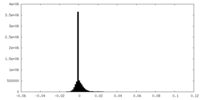
| Density Histograms |
-Half map: HIV-1 CA-SP1 subtomogram average, from the full dataset,...
| File | emd_16209_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | HIV-1 CA-SP1 subtomogram average, from the full dataset, relion4.0 tomo, half2 map | ||||||||||||
| Projections & Slices |
| ||||||||||||
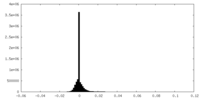
| Density Histograms |
- Sample components
Sample components
-Entire : Human immunodeficiency virus 1
| Entire | Name:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
|---|---|
| Components |
|
-Supramolecule #1: Human immunodeficiency virus 1
| Supramolecule | Name: Human immunodeficiency virus 1 / type: virus / ID: 1 / Parent: 0 Details: HIV-1 dMACANC virus-like particles assembled in the presence of BVM. NCBI-ID: 11676 / Sci species name: Human immunodeficiency virus 1 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: Yes |
|---|---|
| Molecular weight | Theoretical: 74 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Average electron dose: 3.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)