+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Expanded Coxsackievirus A9 after 0.01% faf-BSA treatment | |||||||||||||||
 Map data Map data | Expanded RNA containing CVA9 after 0.01% faf-BSA treatment | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | icosahedral symmetry / expanded virion / picornavirus / A-particle / VIRUS | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport ...symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of RIG-I activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / nucleoside-triphosphate phosphatase / channel activity / monoatomic ion transmembrane transport / DNA replication / RNA helicase activity / endocytosis involved in viral entry into host cell / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / ATP hydrolysis activity / proteolysis / RNA binding / zinc ion binding / ATP binding / membrane Similarity search - Function | |||||||||||||||
| Biological species |  Human coxsackievirus A9 (strain Griggs) / Human coxsackievirus A9 (strain Griggs) /  Coxsackievirus A9 Coxsackievirus A9 | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | |||||||||||||||
 Authors Authors | Domanska A / Plavec Z / Ruokolainen V / Loflund B / Marjomaki VS / Butcher SJ | |||||||||||||||
| Funding support |  Finland, 4 items Finland, 4 items
| |||||||||||||||
 Citation Citation |  Journal: J Virol / Year: 2022 Journal: J Virol / Year: 2022Title: Structural Studies Reveal that Endosomal Cations Promote Formation of Infectious Coxsackievirus A9 A-Particles, Facilitating RNA and VP4 Release. Authors: Aušra Domanska / Zlatka Plavec / Visa Ruokolainen / Benita Löflund / Varpu Marjomäki / Sarah J Butcher /  Abstract: Coxsackievirus A9 (CVA9), an enterovirus, is a common cause of pediatric aseptic meningitis and neonatal sepsis. During cell entry, enterovirus capsids undergo conformational changes leading to ...Coxsackievirus A9 (CVA9), an enterovirus, is a common cause of pediatric aseptic meningitis and neonatal sepsis. During cell entry, enterovirus capsids undergo conformational changes leading to expansion, formation of large pores, externalization of VP1 N termini, and loss of the lipid factor from VP1. Factors such as receptor binding, heat, and acidic pH can trigger capsid expansion in some enteroviruses. Here, we show that fatty acid-free bovine serum albumin or neutral endosomal ionic conditions can independently prime CVA9 for expansion and genome release. Our results showed that CVA9 treatment with albumin or endosomal ions generated a heterogeneous population of virions, which could be physically separated by asymmetric flow field flow fractionation and computationally by cryo-electron microscopy (cryo-EM) and image processing. We report cryo-EM structures of CVA9 A-particles obtained by albumin or endosomal ion treatment and a control nonexpanded virion to 3.5, 3.3, and 2.9 Å resolution, respectively. Whereas albumin promoted stable expanded virions, the endosomal ionic concentrations induced unstable CVA9 virions which easily disintegrated, losing their genome. Loss of most of the VP4 molecules and exposure of negatively charged amino acid residues in the capsid's interior after expansion created a repulsive viral RNA-capsid interface, aiding genome release. Coxsackievirus A9 (CVA9) is a common cause of meningitis and neonatal sepsis. The triggers and mode of action of RNA release into the cell unusually do not require receptor interaction. Rather, a slow process in the endosome, independent of low pH, is required. Here, we show by biophysical separation, cryogenic electron microscopy, and image reconstruction that albumin and buffers mimicking the endosomal ion composition can separately and together expand and prime CVA9 for uncoating. Furthermore, we show in these expanded particles that VP4 is present at only ~10% of the occupancy found in the virion, VP1 is externalized, and the genome is repelled by the negatively charged, repulsive inner surface of the capsid that occurs due to the expansion. Thus, we can now link observations from cell biology of infection with the physical processes that occur in the capsid to promote genome uncoating. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_15692.map.gz emd_15692.map.gz | 80.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-15692-v30.xml emd-15692-v30.xml emd-15692.xml emd-15692.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_15692.png emd_15692.png | 170.1 KB | ||
| Filedesc metadata |  emd-15692.cif.gz emd-15692.cif.gz | 6.4 KB | ||
| Others |  emd_15692_half_map_1.map.gz emd_15692_half_map_1.map.gz emd_15692_half_map_2.map.gz emd_15692_half_map_2.map.gz | 192.4 MB 192.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-15692 http://ftp.pdbj.org/pub/emdb/structures/EMD-15692 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15692 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-15692 | HTTPS FTP |
-Related structure data
| Related structure data |  8aw6MC  8at5C  8axxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_15692.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_15692.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Expanded RNA containing CVA9 after 0.01% faf-BSA treatment | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.97 Å | ||||||||||||||||||||||||||||||||||||
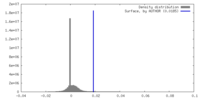
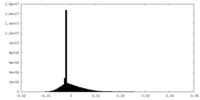
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half map 2 of expanded CVA9 after 0.01% faf-BSA treatment
| File | emd_15692_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 of expanded CVA9 after 0.01% faf-BSA treatment | ||||||||||||
| Projections & Slices |
| ||||||||||||
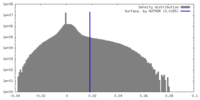
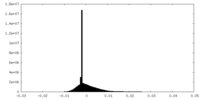
| Density Histograms |
-Half map: Half map 1 of expanded CVA9 after 0.01% faf-BSA treatment
| File | emd_15692_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 of expanded CVA9 after 0.01% faf-BSA treatment | ||||||||||||
| Projections & Slices |
| ||||||||||||
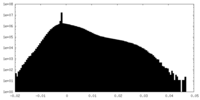
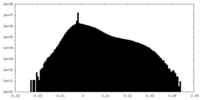
| Density Histograms |
- Sample components
Sample components
-Entire : Coxsackievirus A9
| Entire | Name:  Coxsackievirus A9 Coxsackievirus A9 |
|---|---|
| Components |
|
-Supramolecule #1: Coxsackievirus A9
| Supramolecule | Name: Coxsackievirus A9 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 12067 / Sci species name: Coxsackievirus A9 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8 MDa |
| Virus shell | Shell ID: 1 / Name: icosahedral / Diameter: 300.0 Å / T number (triangulation number): 1 |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Details: Viral protein VP1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human coxsackievirus A9 (strain Griggs) / Strain: Griggs Human coxsackievirus A9 (strain Griggs) / Strain: Griggs |
| Molecular weight | Theoretical: 33.86902 KDa |
| Sequence | String: GDVEEAIERA VVHVADTMRS GPSNSASVPA LTAVETGHTS QVTPSDTMQT RHVKNYHSRS ESTVENFLGR SACVYMEEYK TTDNDVNKK FVAWPINTKQ MVQMRRKLEM FTYLRFDMEV TFVITSRQDP GTTLAQDMPV LTHQIMYVPP GGPIPAKVDD Y AWQTSTNP ...String: GDVEEAIERA VVHVADTMRS GPSNSASVPA LTAVETGHTS QVTPSDTMQT RHVKNYHSRS ESTVENFLGR SACVYMEEYK TTDNDVNKK FVAWPINTKQ MVQMRRKLEM FTYLRFDMEV TFVITSRQDP GTTLAQDMPV LTHQIMYVPP GGPIPAKVDD Y AWQTSTNP SIFWTEGNAP ARMSIPFISI GNAYSNFYDG WSNFDQRGSY GYNTLNNLGH IYVRHVSGSS PHPITSTIRV YF KPKHTRA WVPRPPRLCQ YKKAFSVDFT PTPITDTRKD INTVTTVAQS RRRGDMSTLN TH UniProtKB: Genome polyprotein |
-Macromolecule #2: Capsid protein VP2
| Macromolecule | Name: Capsid protein VP2 / type: protein_or_peptide / ID: 2 / Details: Viral protein VP2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human coxsackievirus A9 (strain Griggs) / Strain: Griggs Human coxsackievirus A9 (strain Griggs) / Strain: Griggs |
| Molecular weight | Theoretical: 28.885518 KDa |
| Sequence | String: SPTVEECGYS DRVRSITLGN STITTQECAN VVVGYGRWPT YLRDDEATAE DQPTQPDVAT CRFYTLDSIK WEKGSVGWWW KFPEALSDM GLFGQNMQYH YLGRAGYTIH VQCNASKFHQ GCLLVVCVPE AEMGGAVVGQ AFSATAMANG DKAYEFTSAT Q SDQTKVQT ...String: SPTVEECGYS DRVRSITLGN STITTQECAN VVVGYGRWPT YLRDDEATAE DQPTQPDVAT CRFYTLDSIK WEKGSVGWWW KFPEALSDM GLFGQNMQYH YLGRAGYTIH VQCNASKFHQ GCLLVVCVPE AEMGGAVVGQ AFSATAMANG DKAYEFTSAT Q SDQTKVQT AIHNAGMGVG VGNLTIYPHQ WINLRTNNSA TIVMPYINSV PMDNMFRHYN FTLMVIPFVK LDYADTASTY VP ITVTVAP MCAEYNGLRL AQAQ UniProtKB: Genome polyprotein |
-Macromolecule #3: Capsid protein VP3
| Macromolecule | Name: Capsid protein VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human coxsackievirus A9 (strain Griggs) / Strain: Griggs Human coxsackievirus A9 (strain Griggs) / Strain: Griggs |
| Molecular weight | Theoretical: 26.335072 KDa |
| Sequence | String: GLPTMNTPGS TQFLTSDDFQ SPCALPQFDV TPSMNIPGEV KNLMEIAEVD SVVPVNNVQD TTDQMEMFRI PVTINAPLQQ QVFGLRLQP GLDSVFKHTL LGEILNYYAH WSGSMKLTFV FCGSAMATGK FLIAYSPPGA NPPKTRKDAM LGTHIIWDIG L QSSCVLCV ...String: GLPTMNTPGS TQFLTSDDFQ SPCALPQFDV TPSMNIPGEV KNLMEIAEVD SVVPVNNVQD TTDQMEMFRI PVTINAPLQQ QVFGLRLQP GLDSVFKHTL LGEILNYYAH WSGSMKLTFV FCGSAMATGK FLIAYSPPGA NPPKTRKDAM LGTHIIWDIG L QSSCVLCV PWISQTHYRL VQQDEYTSAG YVTCWYQTGM IVPPGTPNSS SIMCFASACN DFSVRMLRDT PFISQDNKLQ UniProtKB: Genome polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.1 mg/mL |
|---|---|
| Buffer | pH: 7.25 Details: DPBS containing 0.01% faf-BSA, pH 7.25, incubated at 37 C for 1 h |
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
| Details | this sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: FEI FALCON III (4k x 4k) / Detector mode: COUNTING / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 150000 |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)