+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tail tip of siphophage T5 : full structure | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bacteriophage / Siphophage / T5 / baseplate / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpeptidoglycan muralytic activity / : / symbiont entry into host cell via disruption of host cell wall peptidoglycan / virus tail, tube / symbiont genome ejection through host cell envelope, long flexible tail mechanism / virus tail, baseplate / virus tail, fiber / viral tail assembly / symbiont entry into host cell via disruption of host cell envelope / symbiont entry into host ...peptidoglycan muralytic activity / : / symbiont entry into host cell via disruption of host cell wall peptidoglycan / virus tail, tube / symbiont genome ejection through host cell envelope, long flexible tail mechanism / virus tail, baseplate / virus tail, fiber / viral tail assembly / symbiont entry into host cell via disruption of host cell envelope / symbiont entry into host / virus tail / viral release from host cell by cytolysis / symbiont genome entry into host cell via pore formation in plasma membrane / killing of cells of another organism / lyase activity / defense response to bacterium Similarity search - Function | |||||||||
| Biological species |  Escherichia phage T5 (virus) / Escherichia phage T5 (virus) /  Escherichia virus T5 Escherichia virus T5 | |||||||||
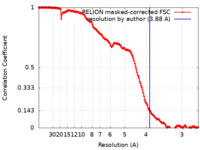
| Method | single particle reconstruction / cryo EM / Resolution: 3.88 Å | |||||||||
 Authors Authors | Linares R / Arnaud CA / Effantin G / Darnault C / Epalle N / Boeri Erba E / Schoehn G / Breyton C | |||||||||
| Funding support |  France, 2 items France, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Structural basis of bacteriophage T5 infection trigger and cell wall perforation. Authors: Romain Linares / Charles-Adrien Arnaud / Grégory Effantin / Claudine Darnault / Nathan Hugo Epalle / Elisabetta Boeri Erba / Guy Schoehn / Cécile Breyton /  Abstract: Most bacteriophages present a tail allowing host recognition, cell wall perforation, and viral DNA channeling from the capsid to the infected bacterium cytoplasm. The majority of tailed phages bear a ...Most bacteriophages present a tail allowing host recognition, cell wall perforation, and viral DNA channeling from the capsid to the infected bacterium cytoplasm. The majority of tailed phages bear a long flexible tail () at the tip of which receptor binding proteins (RBPs) specifically interact with their host, triggering infection. In siphophage T5, the unique RBP is located at the extremity of a central fiber. We present the structures of T5 tail tip, determined by cryo-electron microscopy before and after interaction with its receptor, FhuA, reconstituted into nanodisc. These structures bring out the important conformational changes undergone by T5 tail tip upon infection, which include bending of T5 central fiber on the side of the tail tip, tail anchoring to the membrane, tail tube opening, and formation of a transmembrane channel. The data allow to detail the first steps of an otherwise undescribed infection mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14869.map.gz emd_14869.map.gz | 26.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14869-v30.xml emd-14869-v30.xml emd-14869.xml emd-14869.xml | 29.4 KB 29.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14869_fsc.xml emd_14869_fsc.xml | 16.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_14869.png emd_14869.png | 50.6 KB | ||
| Filedesc metadata |  emd-14869.cif.gz emd-14869.cif.gz | 9.2 KB | ||
| Others |  emd_14869_additional_1.map.gz emd_14869_additional_1.map.gz emd_14869_half_map_1.map.gz emd_14869_half_map_1.map.gz emd_14869_half_map_2.map.gz emd_14869_half_map_2.map.gz | 297.4 MB 298.4 MB 297.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14869 http://ftp.pdbj.org/pub/emdb/structures/EMD-14869 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14869 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14869 | HTTPS FTP |
-Validation report
| Summary document |  emd_14869_validation.pdf.gz emd_14869_validation.pdf.gz | 781.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14869_full_validation.pdf.gz emd_14869_full_validation.pdf.gz | 780.8 KB | Display | |
| Data in XML |  emd_14869_validation.xml.gz emd_14869_validation.xml.gz | 24.1 KB | Display | |
| Data in CIF |  emd_14869_validation.cif.gz emd_14869_validation.cif.gz | 32.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14869 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14869 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14869 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14869 | HTTPS FTP |
-Related structure data
| Related structure data |  7zqbMC  7qg9C  7zhjC  7zlvC  7zn2C  7zn4C  7zqpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_14869.map.gz / Format: CCP4 / Size: 371.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14869.map.gz / Format: CCP4 / Size: 371.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.351 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: unsharpened map
| File | emd_14869_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_14869_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_14869_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia virus T5
| Entire | Name:  Escherichia virus T5 Escherichia virus T5 |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia virus T5
| Supramolecule | Name: Escherichia virus T5 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Pure T5 tails obtained by infecting E. coli F strain with the amber mutant phage T5D20am30d NCBI-ID: 2695836 / Sci species name: Escherichia virus T5 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
-Macromolecule #1: Probable central straight fiber
| Macromolecule | Name: Probable central straight fiber / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
| Molecular weight | Theoretical: 74.851703 KDa |
| Sequence | String: MISNNAPAKM VLNSVLTGYT LAYIQHSIYS DYDVIGRSFW LKEGSNVTRR DFTGIDTFSV TINNLKPTTT YEVQGAFYDS IIDSELLNA QIGINLSDKQ TFKMKSAPRI TGARCESEPV DVGVGAPIVY IDTTGEADYC TIELKDNSNA NNPWVKYYVG A LMPTIMFG ...String: MISNNAPAKM VLNSVLTGYT LAYIQHSIYS DYDVIGRSFW LKEGSNVTRR DFTGIDTFSV TINNLKPTTT YEVQGAFYDS IIDSELLNA QIGINLSDKQ TFKMKSAPRI TGARCESEPV DVGVGAPIVY IDTTGEADYC TIELKDNSNA NNPWVKYYVG A LMPTIMFG GVPIGSYKVR ISGQISLPDG VTIDSSGYYE YPNVFEVRYN FVPPAAPINI VFKAARIADG KERYDLRVQW DW NRGAGAN VREFVLSYID SAEFVRTGWT KAQKINVGAA QSATIISFPW KVEHKFKVSS IAWGPDAQDV TDSAVQTFIL NES TPLDNS FVNETGIEVN YAYIKGKIKD GSTWKQTFLI DAATGAINIG LLDAEGKAPI SFDPVKKIVN VDGSVITKTI NAAN FVMTN LTGQDNPAIY TQGKTWGDTK SGIWMGMDNV TAKPKLDIGN ATQYIRYDGN ILRISSEVVI GTPNGDIDIQ TGIQG KQTV FIYIIGTSLP AKPTSPAYPP SGWSKTPPNR TSNTQNIYCS TGTLDPVTNQ LVSGTSWSDV VQWSGTEGVD GRPGAT GQR GPGMYSLAIA NLTAWNDSQA NSFFTSNFGS GPVKYDVLTE YKSGAPGTAF TRQWNGSAWT SPAMVLHGDM IVNGTVT AS KIVANNAFLS QIGVNIIYDR AAALSSNPEG SYKMKIDLQN GYIHIR UniProtKB: Straight fiber protein pb4 |
-Macromolecule #2: L-shaped tail fiber protein p132
| Macromolecule | Name: L-shaped tail fiber protein p132 / type: protein_or_peptide / ID: 2 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
| Molecular weight | Theoretical: 15.079864 KDa |
| Sequence | String: MSTENRVIDL VVDENVPYGL LMQFMDVDDS VYPSTSKPVD LTDFSLRGSI KSSLEDGAET VASFTTAIVD AAQGVASISL PVSAVTTIA SKASKERDRY NPRQRLAGYY DVIITRTAVG SAASSFRIME GKVYISDGVT Q UniProtKB: Collar protein p132 |
-Macromolecule #3: Minor tail protein
| Macromolecule | Name: Minor tail protein / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
| Molecular weight | Theoretical: 34.367605 KDa |
| Sequence | String: MFYSLMRESK IVIEYDGRGY HFDALSNYDA STSFQEFKTL RRTIHNRTNY ADSIINAQDP SSISLAINFS TTLIESNFFD WMGFTREGN SLFLPRNTPN IEPIMFNMYI INHNNSCIYF ENCYVSTVDF SLDKSIPILN VGIESGKFSE VSTFRDGYTI T QGEVLPYS ...String: MFYSLMRESK IVIEYDGRGY HFDALSNYDA STSFQEFKTL RRTIHNRTNY ADSIINAQDP SSISLAINFS TTLIESNFFD WMGFTREGN SLFLPRNTPN IEPIMFNMYI INHNNSCIYF ENCYVSTVDF SLDKSIPILN VGIESGKFSE VSTFRDGYTI T QGEVLPYS APAVYTNSSP LPALISASMS FQQQCSWRED RNIFDINKIY TNKRAYVNEM NASATLAFYY VKRLVGDKFL NL DPETRTP LIIKNKYVSI TFPLARISKR LNFSDLYQVE YDVIPTADSD PVEINFFGER K UniProtKB: Baseplate tube protein p140 |
-Macromolecule #4: Distal tail protein
| Macromolecule | Name: Distal tail protein / type: protein_or_peptide / ID: 4 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
| Molecular weight | Theoretical: 22.798641 KDa |
| Sequence | String: MRLPDPYTNP EYPGLGFESV NLVDNDPMIR DELPNGKVKE VKISAQYWGI NISYPELFPD EYAFLDSRLL EYKRTGDYLD VLLPQYEAF RVRGDTKSVT IPAGQKGSQI ILNTNGTLTG QPKAGDLFKL STHPKVYKIT NFSSSGNVWN ISLYPDLFIT T TGSEKPVF ...String: MRLPDPYTNP EYPGLGFESV NLVDNDPMIR DELPNGKVKE VKISAQYWGI NISYPELFPD EYAFLDSRLL EYKRTGDYLD VLLPQYEAF RVRGDTKSVT IPAGQKGSQI ILNTNGTLTG QPKAGDLFKL STHPKVYKIT NFSSSGNVWN ISLYPDLFIT T TGSEKPVF NGILFRTKLM NGDSFGSTLN NNGTYSGISL SLRESL UniProtKB: Distal tail protein pb9 |
-Macromolecule #5: Tail tube protein
| Macromolecule | Name: Tail tube protein / type: protein_or_peptide / ID: 5 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
| Molecular weight | Theoretical: 50.459215 KDa |
| Sequence | String: MSLQLLRNTR IFVSTVKTGH NKTNTQEILV QDDISWGQDS NSTDITVNEA GPRPTRGSKR FNDSLNAAEW SFSTYILPYK DKNTSKQIV PDYMLWHALS SGRAINLEGT TGAHNNATNF MVNFKDNSYH ELAMLHIYIL TDKTWSYIDS CQINQAEVNV D IEDIGRVT ...String: MSLQLLRNTR IFVSTVKTGH NKTNTQEILV QDDISWGQDS NSTDITVNEA GPRPTRGSKR FNDSLNAAEW SFSTYILPYK DKNTSKQIV PDYMLWHALS SGRAINLEGT TGAHNNATNF MVNFKDNSYH ELAMLHIYIL TDKTWSYIDS CQINQAEVNV D IEDIGRVT WSGNGNQLIP LDEQPFDPDQ IGIDDETYMT IQGSYIKNKL TILKIKDMDT NKSYDIPITG GTFTINNNIT YL TPNVMSR VTIPIGSFTG AFELTGSLTA YLNDKSLGSM ELYKDLIKTL KVVNRFEIAL VLGGEYDDER PAAILVAKQA HVN IPTIET DDVLGTSVEF KAIPSDLDAG DEGYLGFSSK YTRTTINNLI VNGDGATDAV TAITVKSAGN VTTLNRSATL QMSV EVTPS SARNKEVTWA ITAGDAATIN ATGLLRADAS KTGAVTVEAT AKDGSGVKGT KVITVTAGG UniProtKB: Tail tube protein pb6 |
-Macromolecule #6: Pore-forming tail tip protein pb2
| Macromolecule | Name: Pore-forming tail tip protein pb2 / type: protein_or_peptide / ID: 6 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
| Molecular weight | Theoretical: 131.629547 KDa |
| Sequence | String: MTDKLIRELL IDVKQKGATR TAKSIENVSD ALENAAAASE LTNEQLGKMP RTLYSIERAA DRAAKSLTKM QASRGMAGIT KSIDGIGDK LDYLAIQLIE VTDKLEIGFD GVSRSVKAMG NDVAAATEKV QDRLYDTNRA LGGTSKGFND TAGAAGRASR A LGNTSGSA ...String: MTDKLIRELL IDVKQKGATR TAKSIENVSD ALENAAAASE LTNEQLGKMP RTLYSIERAA DRAAKSLTKM QASRGMAGIT KSIDGIGDK LDYLAIQLIE VTDKLEIGFD GVSRSVKAMG NDVAAATEKV QDRLYDTNRA LGGTSKGFND TAGAAGRASR A LGNTSGSA RGATRDFAAM AKIGGRLPIM YAALASNVFV LQTAFESLKV GDQLNRLEQF GTIVGTMTGT PVQTLALSLQ NA TNGAISF EEAMRQASSA SAYGFDSEQL EQFGLVARRA AAVLGVDMTD ALNRVIKGVS KQEIELLDEL GVTIRLNDAY ENY VKQLNA TSTGIKYTVD SLTTYQKQQA YANEVIAEST RRFGYLDDAL KATSWEQFAA NANSALRSLQ QSAATYLNPV MDTL NTFLY QTKSSQMRVS AMARSASAKT TPAENVTALI ENAVGAREDL DTYLKESEER VKKAQELKQQ LDDLKAKQAA TAPIA NALT AGGIGGDESN KLVVQLTNEL ARQNKEIEER TKTEKVLRQA VQDTGEALLR NGKLAEQLGA KMKYADTAVP GDKGVF EVD PNNLKAVSEI QKNFDFLKKS SSDTANNIRM AASSITNAKK ASSDLNSVVK AVEDTSKVTG QSADTLVKNL NLGFSSL DQ MKAAQKGLSE YVTAMDKSEQ NALEVAKRKD EVYNQTKDKA KAEAAAREVL LRQQQEQLTA AKALLAINPN DPEALKQV A KIETEILNTK AQGFENAKKT KDYTDKILGV DREIALLNDR TMTSTQYRLA QLRLELQLEQ EKTELYSKQA DGQAKVEQS RRAQAQISRE IWEAEKQGTA SHVSALMDAL EVSQTQRNVT GQSQILTERL SILQQQLELS KGNTEEELKY RNEIYKTSAA LEQLKKQRE SQMQQQVGSS VGATYTSTTG LIGEDKDFAD MQNRMASYDQ AISKLSELNS EATAVAQSMG NLTNAMIQFS Q GSLDTTSM IASGMQTVAS MIQYSTSQQV SAIDQAIAAE QKRDGKSEAS KAKLKKLEAE KLKIQQDAAK KQIIIQTAVA VM QAATAVP YPFSIPLMVA AGLAGALALA QASSASGMSS IADSGADTTQ YLTLGERQKN VDVSMQASSG ELSYLRGDKG IGN ANSFVP RAEGGMMYPG VSYQMGEHGT EVVTPMVPMK ATPNDQLSDG SKTTSGRPII LNISTMDAAS FRDFASNNST AFRD AVELA LNENGTTLKS LGNS UniProtKB: Tape measure protein pb2 precursor |
-Macromolecule #7: Probable baseplate hub protein
| Macromolecule | Name: Probable baseplate hub protein / type: protein_or_peptide / ID: 7 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
| Molecular weight | Theoretical: 107.279766 KDa |
| Sequence | String: MKKILDSAKN YLNTHDKLKT ACLIALELPS SSGSAATYIY LTDYFRDVTY NGILYRSGKV KSISSHKQNR QLSIGSLSFT ITGTAEDEV LKLVQNGVSF LDRGITIHQA IINEEGNILP VDPDTDGPLL FFRGRITGGG IKDNVNTSGI GTSVITWNCS N QFYDFDRV ...String: MKKILDSAKN YLNTHDKLKT ACLIALELPS SSGSAATYIY LTDYFRDVTY NGILYRSGKV KSISSHKQNR QLSIGSLSFT ITGTAEDEV LKLVQNGVSF LDRGITIHQA IINEEGNILP VDPDTDGPLL FFRGRITGGG IKDNVNTSGI GTSVITWNCS N QFYDFDRV NGRYTDDASH RGLEVVNGTL QPSNGAKRPE YQEDYGFFHS NKSTTILAKY QVKEERYKLQ SKKKLFGLSR SY SLKKYYE TVTKEVDLDF NLAAKFIPVV YGVQKIPGIP IFADTELNNP NIVYVVYAFA EGEIDGFLDF YIGDSPMICF DET DSDTRT CFGRKKIVGD TMHRLAAGTS TSQPSVHGQE YKYNDGNGDI RIWTFHGKPD QTAAQVLVDI AKKKGFYLQN QNGN GPEYW DSRYKLLDTA YAIVRFTINE NRTEIPEISA EVQGKKVKVY NSDGTIKADK TSLNGIWQLM DYLTSDRYGA DITLD QFPL QKVISEAKIL DIIDESYQTS WQPYWRYVGW NDPLSENRQI VQLNTILDTS ESVFKNVQGI LESFGGAINN LSGEYR ITV EKYSTNPLRI NFLDTYGDLD LSDTTGRNKF NSVQASLVDP ALSWKTNSIT FYNSKFKEQD KGLDKKLQLS FANITNY YT ARSYADRELK KSRYSRTLSF SVPYKFIGIE PNDPIAFTYE RYGWKDKFFL VDEVENTRDG KINLVLQEYG EDVFINSE Q VDNSGNDIPD ISNNVLPPRD FKYTPTPGGV VGAIGKNGEL SWLPSLTNNV VYYSIAHSGH VNPYIVQQLE NNPNERMIQ EIIGEPAGLA IFELRAVDIN GRRSSPVTLS VDLNSAKNLS VVSNFRVVNT ASGDVTEFVG PDVKLAWDKI PEEEIIPEIY YTLEIYDSQ DRMLRSVRIE DVYTYDYLLT YNKADFALLN SGALGINRKL RFRIRAEGEN GEQSVGWATI UniProtKB: Baseplate hub protein pb3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER/RHODIUM / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Details: Intensity 25 mA | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293.15 K / Instrument: FEI VITROBOT MARK IV Details: 3 uL of T5 tails sample (with or without FhuA-ND) were deposited on a freshly glow discharged EM grid and plunge-frozen in nitrogen-cooled liquid ethane using a ThermoFisher Mark IV Vitrobot ...Details: 3 uL of T5 tails sample (with or without FhuA-ND) were deposited on a freshly glow discharged EM grid and plunge-frozen in nitrogen-cooled liquid ethane using a ThermoFisher Mark IV Vitrobot device (100 percent humidity, 20 Celsius degrees, 5 s blotting time, blot force 0). | |||||||||||||||
| Details | Pure T5 tails obtained by infecting E. coli F strain with the amber mutant phage T5D20am30d |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Atomic protein models were built into the different cryo-EM maps using the Coot software by tracing the protein sequence into the densities and were then iteratively refined alternating Coot manual refinement and PHENIX real space refine tool until convergence. p140, p132, BHPpb3 and TMPpb2 C-ter models were built ab initio. For TTPpb6 and DTPpb9 models, already existing X-ray models (PDB codes 5NGJ / 4JMQ) were used as a starting point and were refined into the EM maps. Molprobity was used for model quality assessment. |
|---|---|
| Refinement | Space: REAL / Protocol: BACKBONE TRACE / Overall B value: 40 |
| Output model |  PDB-7zqb: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)