+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Tail tip of siphophage T5 : common core proteins | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Bacteriophage / Siphophage / T5 / baseplate / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationvirus tail, tube / symbiont genome ejection through host cell envelope, long flexible tail mechanism / virus tail, baseplate / virus tail, fiber / virus tail / viral release from host cell by cytolysis Similarity search - Function | |||||||||
| Biological species |  Escherichia phage T5 (virus) / Escherichia phage T5 (virus) /  Escherichia virus T5 Escherichia virus T5 | |||||||||
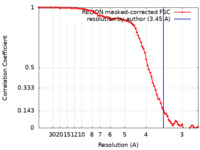
| Method | single particle reconstruction / cryo EM / Resolution: 3.45 Å | |||||||||
 Authors Authors | Linares R / Arnaud CA / Effantin G / Darnault C / Epalle N / Boeri Erba E / Schoehn G / Breyton C | |||||||||
| Funding support |  France, 2 items France, 2 items
| |||||||||
 Citation Citation |  Journal: Sci Adv / Year: 2023 Journal: Sci Adv / Year: 2023Title: Structural basis of bacteriophage T5 infection trigger and cell wall perforation. Authors: Romain Linares / Charles-Adrien Arnaud / Grégory Effantin / Claudine Darnault / Nathan Hugo Epalle / Elisabetta Boeri Erba / Guy Schoehn / Cécile Breyton /  Abstract: Most bacteriophages present a tail allowing host recognition, cell wall perforation, and viral DNA channeling from the capsid to the infected bacterium cytoplasm. The majority of tailed phages bear a ...Most bacteriophages present a tail allowing host recognition, cell wall perforation, and viral DNA channeling from the capsid to the infected bacterium cytoplasm. The majority of tailed phages bear a long flexible tail () at the tip of which receptor binding proteins (RBPs) specifically interact with their host, triggering infection. In siphophage T5, the unique RBP is located at the extremity of a central fiber. We present the structures of T5 tail tip, determined by cryo-electron microscopy before and after interaction with its receptor, FhuA, reconstituted into nanodisc. These structures bring out the important conformational changes undergone by T5 tail tip upon infection, which include bending of T5 central fiber on the side of the tail tip, tail anchoring to the membrane, tail tube opening, and formation of a transmembrane channel. The data allow to detail the first steps of an otherwise undescribed infection mechanism. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_13953.map.gz emd_13953.map.gz | 7.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-13953-v30.xml emd-13953-v30.xml emd-13953.xml emd-13953.xml | 22.1 KB 22.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_13953_fsc.xml emd_13953_fsc.xml | 8.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_13953.png emd_13953.png | 133.2 KB | ||
| Masks |  emd_13953_msk_1.map emd_13953_msk_1.map | 52.7 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-13953.cif.gz emd-13953.cif.gz | 7.2 KB | ||
| Others |  emd_13953_half_map_1.map.gz emd_13953_half_map_1.map.gz emd_13953_half_map_2.map.gz emd_13953_half_map_2.map.gz | 40.8 MB 40.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-13953 http://ftp.pdbj.org/pub/emdb/structures/EMD-13953 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13953 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-13953 | HTTPS FTP |
-Related structure data
| Related structure data |  7qg9MC  7zhjC  7zlvC  7zn2C  7zn4C  7zqbC  7zqpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_13953.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_13953.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.351 Å | ||||||||||||||||||||||||||||||||||||
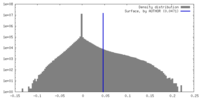
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_13953_msk_1.map emd_13953_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_13953_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_13953_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Escherichia virus T5
| Entire | Name:  Escherichia virus T5 Escherichia virus T5 |
|---|---|
| Components |
|
-Supramolecule #1: Escherichia virus T5
| Supramolecule | Name: Escherichia virus T5 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Pure T5 tails obtained by infecting E. coli F strain with the amber mutant phage T5D20am30d, incubated or not with the bacterial receptor FhuA NCBI-ID: 2695836 / Sci species name: Escherichia virus T5 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
-Macromolecule #1: L-shaped tail fiber protein p132
| Macromolecule | Name: L-shaped tail fiber protein p132 / type: protein_or_peptide / ID: 1 / Number of copies: 12 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
| Molecular weight | Theoretical: 15.079864 KDa |
| Sequence | String: MSTENRVIDL VVDENVPYGL LMQFMDVDDS VYPSTSKPVD LTDFSLRGSI KSSLEDGAET VASFTTAIVD AAQGVASISL PVSAVTTIA SKASKERDRY NPRQRLAGYY DVIITRTAVG SAASSFRIME GKVYISDGVT Q UniProtKB: Collar protein p132 |
-Macromolecule #2: Minor tail protein
| Macromolecule | Name: Minor tail protein / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
| Molecular weight | Theoretical: 34.367605 KDa |
| Sequence | String: MFYSLMRESK IVIEYDGRGY HFDALSNYDA STSFQEFKTL RRTIHNRTNY ADSIINAQDP SSISLAINFS TTLIESNFFD WMGFTREGN SLFLPRNTPN IEPIMFNMYI INHNNSCIYF ENCYVSTVDF SLDKSIPILN VGIESGKFSE VSTFRDGYTI T QGEVLPYS ...String: MFYSLMRESK IVIEYDGRGY HFDALSNYDA STSFQEFKTL RRTIHNRTNY ADSIINAQDP SSISLAINFS TTLIESNFFD WMGFTREGN SLFLPRNTPN IEPIMFNMYI INHNNSCIYF ENCYVSTVDF SLDKSIPILN VGIESGKFSE VSTFRDGYTI T QGEVLPYS APAVYTNSSP LPALISASMS FQQQCSWRED RNIFDINKIY TNKRAYVNEM NASATLAFYY VKRLVGDKFL NL DPETRTP LIIKNKYVSI TFPLARISKR LNFSDLYQVE YDVIPTADSD PVEINFFGER K UniProtKB: Baseplate tube protein p140 |
-Macromolecule #3: Distal tail protein
| Macromolecule | Name: Distal tail protein / type: protein_or_peptide / ID: 3 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
| Molecular weight | Theoretical: 22.798641 KDa |
| Sequence | String: MRLPDPYTNP EYPGLGFESV NLVDNDPMIR DELPNGKVKE VKISAQYWGI NISYPELFPD EYAFLDSRLL EYKRTGDYLD VLLPQYEAF RVRGDTKSVT IPAGQKGSQI ILNTNGTLTG QPKAGDLFKL STHPKVYKIT NFSSSGNVWN ISLYPDLFIT T TGSEKPVF ...String: MRLPDPYTNP EYPGLGFESV NLVDNDPMIR DELPNGKVKE VKISAQYWGI NISYPELFPD EYAFLDSRLL EYKRTGDYLD VLLPQYEAF RVRGDTKSVT IPAGQKGSQI ILNTNGTLTG QPKAGDLFKL STHPKVYKIT NFSSSGNVWN ISLYPDLFIT T TGSEKPVF NGILFRTKLM NGDSFGSTLN NNGTYSGISL SLRESL UniProtKB: Distal tail protein pb9 |
-Macromolecule #4: Tail tube protein
| Macromolecule | Name: Tail tube protein / type: protein_or_peptide / ID: 4 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage T5 (virus) Escherichia phage T5 (virus) |
| Molecular weight | Theoretical: 50.459215 KDa |
| Sequence | String: MSLQLLRNTR IFVSTVKTGH NKTNTQEILV QDDISWGQDS NSTDITVNEA GPRPTRGSKR FNDSLNAAEW SFSTYILPYK DKNTSKQIV PDYMLWHALS SGRAINLEGT TGAHNNATNF MVNFKDNSYH ELAMLHIYIL TDKTWSYIDS CQINQAEVNV D IEDIGRVT ...String: MSLQLLRNTR IFVSTVKTGH NKTNTQEILV QDDISWGQDS NSTDITVNEA GPRPTRGSKR FNDSLNAAEW SFSTYILPYK DKNTSKQIV PDYMLWHALS SGRAINLEGT TGAHNNATNF MVNFKDNSYH ELAMLHIYIL TDKTWSYIDS CQINQAEVNV D IEDIGRVT WSGNGNQLIP LDEQPFDPDQ IGIDDETYMT IQGSYIKNKL TILKIKDMDT NKSYDIPITG GTFTINNNIT YL TPNVMSR VTIPIGSFTG AFELTGSLTA YLNDKSLGSM ELYKDLIKTL KVVNRFEIAL VLGGEYDDER PAAILVAKQA HVN IPTIET DDVLGTSVEF KAIPSDLDAG DEGYLGFSSK YTRTTINNLI VNGDGATDAV TAITVKSAGN VTTLNRSATL QMSV EVTPS SARNKEVTWA ITAGDAATIN ATGLLRADAS KTGAVTVEAT AKDGSGVKGT KVITVTAGG UniProtKB: Tail tube protein pb6 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER/RHODIUM / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Details: intensity 25 mA | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293.15 K / Instrument: FEI VITROBOT MARK IV Details: 3 uL of T5 tails sample (with or without FhuA-ND) were deposited on a freshly glow discharged EM grid and plunge-frozen in nitrogen-cooled liquid ethane using a ThermoFisher Mark IV Vitrobot ...Details: 3 uL of T5 tails sample (with or without FhuA-ND) were deposited on a freshly glow discharged EM grid and plunge-frozen in nitrogen-cooled liquid ethane using a ThermoFisher Mark IV Vitrobot device (100 percent humidity, 20 Celsius degrees, 5 s blotting time, blot force 0). | |||||||||||||||
| Details | Pure T5 tails obtained by infecting E. coli F strain with the amber mutant phage T5D20am30d, incubated or not with the bacterial receptor FhuA |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | Atomic protein models were built into the different cryo-EM maps using the Coot software by tracing the protein sequence into the densities and were then iteratively refined alternating Coot manual refinement and PHENIX real space refine tool until convergence. p140, p132, BHPpb3 and TMPpb2 C-ter models were built ab initio. For TTPpb6 and DTPpb9 models, already existing X-ray models (PDB codes 5NGJ / 4JMQ) were used as a starting point and were refined into the EM maps. Molprobity was used for model quality assessment. |
|---|---|
| Refinement | Space: REAL / Protocol: BACKBONE TRACE / Overall B value: 112 |
| Output model |  PDB-7qg9: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)