+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mouse RNF213, with mixed nucleotides bound | |||||||||||||||
 Map data Map data | full non-sharpened map | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | nucleotide / AAA / RNF213 / E3 ligase / SIGNALING PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationlipid ubiquitination / negative regulation of non-canonical Wnt signaling pathway / lipid droplet formation / xenophagy / Antigen processing: Ubiquitination & Proteasome degradation / sprouting angiogenesis / Transferases; Acyltransferases; Aminoacyltransferases / regulation of lipid metabolic process / protein K63-linked ubiquitination / immune system process ...lipid ubiquitination / negative regulation of non-canonical Wnt signaling pathway / lipid droplet formation / xenophagy / Antigen processing: Ubiquitination & Proteasome degradation / sprouting angiogenesis / Transferases; Acyltransferases; Aminoacyltransferases / regulation of lipid metabolic process / protein K63-linked ubiquitination / immune system process / protein autoubiquitination / lipid droplet / RING-type E3 ubiquitin transferase / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / ubiquitin-protein transferase activity / ubiquitin protein ligase activity / angiogenesis / ubiquitin-dependent protein catabolic process / defense response to bacterium / protein ubiquitination / ATP hydrolysis activity / zinc ion binding / ATP binding / cytoplasm / cytosol Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
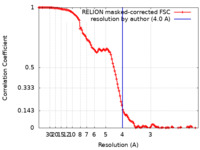
| Method | single particle reconstruction / cryo EM / Resolution: 4.0 Å | |||||||||||||||
 Authors Authors | Ahel J / Clausen T | |||||||||||||||
| Funding support |  Austria, Austria,  United Kingdom, 4 items United Kingdom, 4 items
| |||||||||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: E3 ubiquitin ligase RNF213 employs a non-canonical zinc finger active site and is allosterically regulated by ATP Authors: Ahel J / Fletcher AJ / Grabarczyk D / Roitinger E / Deszcz L / Lehner A / Virdee S / Clausen T | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12932.map.gz emd_12932.map.gz | 131.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12932-v30.xml emd-12932-v30.xml emd-12932.xml emd-12932.xml | 34.9 KB 34.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12932_fsc.xml emd_12932_fsc.xml | 12.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_12932.png emd_12932.png | 139.4 KB | ||
| Masks |  emd_12932_msk_1.map emd_12932_msk_1.map emd_12932_msk_2.map emd_12932_msk_2.map | 166.4 MB 166.4 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-12932.cif.gz emd-12932.cif.gz | 10 KB | ||
| Others |  emd_12932_additional_1.map.gz emd_12932_additional_1.map.gz emd_12932_additional_2.map.gz emd_12932_additional_2.map.gz emd_12932_additional_3.map.gz emd_12932_additional_3.map.gz emd_12932_additional_4.map.gz emd_12932_additional_4.map.gz emd_12932_additional_5.map.gz emd_12932_additional_5.map.gz emd_12932_half_map_1.map.gz emd_12932_half_map_1.map.gz emd_12932_half_map_2.map.gz emd_12932_half_map_2.map.gz | 152.8 MB 152.5 MB 131.1 MB 131.5 MB 131.5 MB 131.6 MB 131.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12932 http://ftp.pdbj.org/pub/emdb/structures/EMD-12932 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12932 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12932 | HTTPS FTP |
-Related structure data
| Related structure data |  7oimMC  7oikC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
| EM raw data |  EMPIAR-10712 (Title: Transmission electron micrographs of mouse RNF213 incubated with ATPγS EMPIAR-10712 (Title: Transmission electron micrographs of mouse RNF213 incubated with ATPγSData size: 2.3 TB Data #1: unaligned multi-frame micrographs from a dataset collected without grid tilting [micrographs - multiframe] Data #2: unaligned multi-frame micrographs from a dataset collected with a 30 deg grid tilt [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_12932.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12932.map.gz / Format: CCP4 / Size: 166.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full non-sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.053 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_12932_msk_1.map emd_12932_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
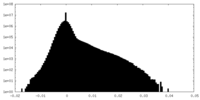
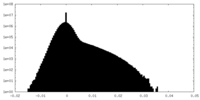
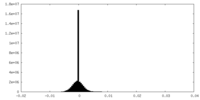
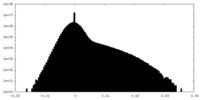
| Density Histograms |
-Mask #2
| File |  emd_12932_msk_2.map emd_12932_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
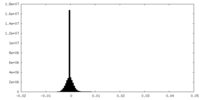
| Density Histograms |
-Additional map: full sharpened map
| File | emd_12932_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full sharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
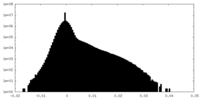
| Density Histograms |
-Additional map: sharpened focused map, focused on AAA core
| File | emd_12932_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened focused map, focused on AAA core | ||||||||||||
| Projections & Slices |
| ||||||||||||
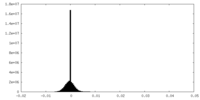
| Density Histograms |
-Additional map: non-sharpened focused map, focused on AAA core
| File | emd_12932_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | non-sharpened focused map, focused on AAA core | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: non-sharpened focused half map 1, focused on AAA cpre
| File | emd_12932_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | non-sharpened focused half map 1, focused on AAA cpre | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: non-sharpened focused half map 2, focused on AAA cpre
| File | emd_12932_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | non-sharpened focused half map 2, focused on AAA cpre | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: full non-sharpened map, half volume 2
| File | emd_12932_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full non-sharpened map, half volume 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: full non-sharpened map, half volume 1
| File | emd_12932_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | full non-sharpened map, half volume 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : RNF213 incubated with ATPgS
| Entire | Name: RNF213 incubated with ATPgS |
|---|---|
| Components |
|
-Supramolecule #1: RNF213 incubated with ATPgS
| Supramolecule | Name: RNF213 incubated with ATPgS / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 580 KDa |
-Macromolecule #1: E3 ubiquitin-protein ligase RNF213
| Macromolecule | Name: E3 ubiquitin-protein ligase RNF213 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO / EC number: RING-type E3 ubiquitin transferase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 552.623375 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASWSHPQFE KGSTSAFNPR DTVTVYFHAI VSRHFGFNPE EHKVYVRGGE GLGQKGWTDA CEMYCTQDLH DLGSLVEGKM DIPRQSLDK PIPYKYVIHR GGSSKDTVEY EFIYEQAQKK GEHVNRCLRV VSTSLGNGDW HQYDDIICMR STGFFQQAKN R ILDSTRKE ...String: MASWSHPQFE KGSTSAFNPR DTVTVYFHAI VSRHFGFNPE EHKVYVRGGE GLGQKGWTDA CEMYCTQDLH DLGSLVEGKM DIPRQSLDK PIPYKYVIHR GGSSKDTVEY EFIYEQAQKK GEHVNRCLRV VSTSLGNGDW HQYDDIICMR STGFFQQAKN R ILDSTRKE LLKGKKQAAV VMLDRIFSVL QPWSDINLQS FMTQFLQFYS VVREPMIHDG RARKWTSLQY EEKEVWTNLW EH VKKQMAP FLEGKSGESL PADCPVRSKL TLGLSILFMV EAAEFTVPKK DLDSLCYLLI PSAGSPEALH SDLSPVLRIR QRW RIYLTN LCLRCIDERC DRWLGILPLL HTCMQKSPPK KNSKSQPEDT WAGLEGISFS EFRDKAPTRS QPLQFMQSKM ALLR VDEYL FRSWLSVVPL ESLSSYLENS IDYLSDVPVR VLDCLQGISY RLPGLRKISN QNMKKDVENV FKMLMHLVDI YQHRI FGEN LLQIYLTECL TLHETVCNIT ANHQFFEIPA LSAELICKLL ELSPPGHTDE GLPEKSYEDL VTSTLQEALA TTRNWL RSL FKSRMLSISS AYVRLTYSEE MAVWRRLVEI GFPEKHGWKG SLLGDMEGRL KQEPPRLQIS FFCSSQCRDG GLHDSVS RS FEKCVIEAVS SACQSQTSVL EGLSCQDLQK FGTLLSAVIT KSWPVHNGEP VFDVDEIFKY LLKWPDVRQL FELCGTNE K IIDNITEEGR QLMATAESVF QKVAGELENG TIVVGQLELI LEHQSQFLDI WNLNRRRLPS QEKACDVRSL LKRRRDDLL FLKQEKRYVE SLLRQLGRVK HLVQVDFGNI EIIHSQDLSN KKLNEAVIKL PNSSSYKRET HYCLSPDIRE MASKLDSLKD SHIFQDFWQ ETAESLNTLD KDPRELKVSL PEVLEYLYNP CYDNFYTLYE NLKSGKITFA EVDAIFKDFV DKYDELKNDL K FMCTMNPQ DQKGWISERV GQIKEYHTLH QAVSSAKVIL QVRRALGVTG DFSVLNPLLN FADSFEDFGN EKLDQISPQF IK AKQLLQD ISEPRQRCLE ELARQTELVA WLHKALEDIN ELKVFVDLAS ISAGENDIDV DRVACFHDAV QGYASLLYKM DER TNFSDF MNHLQELWRA LDNDQHLPDK LKDSARNLEW LKTVKESHGS VELSSLSLAT AINSRGVYVI EAPKDGQKIS PDTV LRLLL PDGHGYPEAL RTYSTEELKE LLNKLMLMSG KKDHNSNTEV EKFSEVFSNM QRLVHVFIKL HCAGNMLFRT WTAKV YCCP DGGIFMNFGL ELLSQLTEKG DVIQLLGALC RQMEDFLDNW KTVVAQKRAE HFYLNFYTAE QLVYLSSELR KPRPSE AAL MMLSFIKGKC TVQDLVQATS ACESKADRYC LREVMKKLPQ QLLSEPSLMG KLQVIMMQSL VYMSAFLPHC LDLDALG RC LAHLATMGGT PVERPLPKGL QAGQPNLILC GHSEVLPAAL AIYMQAPRQP LPTFDEVLLC TPATTIEEVE LLLRRCLT S GSQGHKVYSL LFADQLSYEV GCQAEEFFQS LCTRAHREDY QLVILCDAAR EHCYIPSTFS QYKVPLVPQA PLPNIQAYL QSHYQVPKRL LSAATVFRDG LCVGIVTSER AGVGKSLYVN TLHTKLKAKL RDETVPLKII RLTEPHLDEN QVLSALLPFL KEKYQKMPV IFHIDISTSV QTGIPIFLFK LLILQYLMDI NGKIWRRSPG HLYLVEIPQG LSVQPKRSSK LNARAPLFKF L DLFPKVTC RPPKEVIDME LTPERSHTDP AMDPVEFCSE AFQRPYQYLK RFHQQQNLDT FQYEKGSVEG SPEECLQHFL IY CGLINPS WSELRNFAWF LNCQLKDCEA SIFCKSAFTG DTLRGFKNFV VTFMILMARD FATPTLHTSD QSPGRQSVTI GEV VEEDLA PFSLRKRWES EPHPYVFFNG DHMTMTFIGF HLETNNNGYV DAINPSNGKV IKKDVMTKEL FDGLRLQRVP FNID FDNLP RYEKLERLCL ALGIEWPIDP DETYELTTDN MLKILAIEMR FRCGIPVIIM GETGCGKTRL IKFLSDLKRG SVEAE TMKL VKVHGGTTPS MIYSKVKEAE RTAFSNKAQH KLDTILFFDE ANTTEAVSCI KEILCDRTVD GEHLHEDSGL HIIAAC NPY RKHSQEMILR LESAGLGYRV SAEETADRLG SIPLRQLVYR VHALPPSLIP LVWDFGQLND SAEKLYIQQI VQRLVDS VS VNPSETCVIA DVLSASQMFM RKRENECGFV SLRDVERCVK VFRWFHDHSD MLLKELDKFL HESSDSTHTF ERDPVLWS L VMAIGVCYHA SLEEKASYRT AIARCFPKPY NSSRAILDEV THVQDLFLRG APIRTNIARN LALKENVFMM VICIELKIP LFLVGKPGSS KSLAKIIVAD AMQGQAAFSE LFRCLKQVHL VSFQCSPHST PQGIISTFKQ CARFQQGKDL GQYVSVVVLD EVGLAEDSP KMPLKTLHPL LEDGCIEDDP APYKKVGFVG ISNWALDPAK MNRGIFVSRG SPNEKELIES AEGICSSDRL V QDKIRGYF APFAKAYETV CQKQDKEFFG LRDYYSLIKM VFAKAKASKR GLSPQDITHA VLRNFSGKDN IQALSIFTAS LP EARYKEE VSTVELIKQN IYPGPQASSR GLDGAESRYL LVLTRNYVAL QILQQTFFEG QQPEIIFGSS FPQDQEYTQI CRN INRVKI CMETGKMVVL LNLQNLYESL YDALNQYYVY LGGQKYVDLG LGTHRVKCRV HTAFRLIVIE EKDVVYKQFP VPLI NRLEK HYLDMNTVLQ PWQKSIVQEL QQWAHEFADV KADQFIARHK YSPADVFIGY HSDACASVVL QAVERQGCRD LTEEL YRKV SEEARSILLD CATPDAVVRL SGSSLGSFTA KQLSQEYYYA QQHNSFVDFL QAHLRMTHHE CRAVFTEITT FSRLLT GND CDVLASELRG LASKPVVLSL QQYDTEYSFL KDVRSWLTNP GKRKVLVIQA DFDDGTRSAQ LVASAKYTAI NEINKTQ GT KDFVFVYFVT KLSRMGSGTS YVGFHGGLWR SVHIDDLRRS TIMASDVTKL QNVTISQLFK PEDKPEQEEM EIETSQSK E LAEEQMEVED SEEMKKASDP RSCDCSQFLD TTRLVQSCVQ GAVGMLRDQN ESCARNMRRV TILLDLLNED NTRNASFLR ESKMRLHVLL NKQEENQVRS LKEWVTREAA NQDALQEAGT FRHTLWKRVQ DVVTPILASM IAHIDRDGNL ELLAQPDSPA WVQDLWMFI YSDIKFLNIS LVLNNTRSNS EMSFILVQSH MNLLKDAYNA VPFSWRIRDY LEELWVQAQY ITDTEGLSKK F VEIFQKTP LGVFLAQFPV AQQQKLLQSY LKDFLLLTMK VSSREELMFL QMALWSCLRE LQEASGTPDE TYKFPLSLPW VH LAFQHFR TRLQNFSRIL TIHPQVLSSL SQAAEKHSLA GCEMTLDAFA AMACAEMLKG DLLKPSPKAW LQLVKNLSTP LEL VCSEGY LCDSGSMTRS VIQEVRALWN RIFSIALFVE HVLLGTESHI PELSPLVTTY VSLLDKCLEE DSNLKTCRPF VAVM TTLCD CKDKASKKFS RFGIQPCFIC HGDAQDPVCL PCDHVYCLRC IQTWLIPGQM MCPYCLTDLP DKFSPTVSQD HRKAI EKHA QFRHMCNSFF VDLVSTMCFK DNTPPEKSVI DTLLSLLFVQ KELLRDASQK HREHTKSLSP FDDVVDQTPV IRSVLL KLL LKYSFHEVKD YIQNYLTQLE KKAFLTEDKT ELYLLFISCL EDSVHQKTSA GCRNLEQVLR EEGHFLRTYS PGLQGQE PV RIASVEYLQE VARVRLCLDL AADFLSELQE GSELAEDKRR FLKHVEEFCT RVNNDWHRVY LVRKLSSQRG MEFVQSFS K QGHPCQWVFP RKVIAQQKDH VSLMDRYLVH GNEYKAVRDA TAKAVLECKT LDIGNALMAC RSPKPQQTAY LLLALYTEV AALYRSPNGS LHPEAKQLEA VNKFIKESKI LSDPNIRCFA RSLVDNTLPL LKIRSANSIL KGTVTEMAVH VATILLCGHN QILKPLRNL AFYPVNMANA FLPTMPEDLL VHARTWRGLE NVTWYTCPRG HPCSVGECGR PMQESTCLDC GLPVGGLNHT P HEGFSAIR NNEDRTQTGH VLGSPQSSGV AEVSDRGQSP VVFILTRLLT HLAMLVGATH NPQALTVIIK PWVQDPQGFL QQ HIQRDLE QLTKMLGRSA DETIHVVHLI LSSLLRVQSH GVLNFNAELS TKGCRNNWEK HFETLLLREL KHLDKNLPAI NAL ISQDER ISSNPVTKII YGDPATFLPH LPQKSIIHCS KIWSCRRKIT VEYLQHIVEQ KNGKETVPVL WHFLQKEAEL RLVK FLPEI LALQRDLVKQ FQNVSRVEYS SIRGFIHSHS SDGLRKLLHD RITIFLSTWN ALRRSLETNG EIKLPKDYCC SDLDL DAEF EVILPRRQGL GLCGTALVSY LISLHNNMVY TVQKFSNEDN SYSVDISEVA DLHVISYEVE RDLNPLILSN CQYQVQ QGG ETSQEFDLEK IQRQISSRFL QGKPRLTLKG IPTLVYRRDW NYEHLFMDIK NKMAQSSLPN LAISTISGQL QSYSDAC EA LSIIEITLGF LSTAGGDPGM DLNVYIEEVL RMCDQTAQVL KAFSRCQLRH IIALWQFLSA HKSEQRLRLN KELFREID V QYKEELSTQH QRLLGTFLNE AGLDAFLLEL HEMIVLKLKG PRAANSFNPN WSLKDTLVSY METKDSDILS EVESQFPEE ILMSSCISVW KIAATRKWDR QSRGGGHHHH HHHHHH UniProtKB: E3 ubiquitin-protein ligase RNF213 |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 1 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #5: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #6: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 6 / Number of copies: 1 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 Component:
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: UltrAuFoil R2/2 / Material: GOLD / Mesh: 200 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR | ||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: LEICA PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | #0 - Image recording ID: 1 / #0 - Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / #0 - Number grids imaged: 1 / #0 - Number real images: 3055 / #0 - Average exposure time: 2.0 sec. / #0 - Average electron dose: 45.0 e/Å2 / #0 - Details: grid tilted 30 degrees 1 image per "hole" / #1 - Image recording ID: 2 / #1 - Film or detector model: GATAN K2 QUANTUM (4k x 4k) / #1 - Detector mode: COUNTING / #1 - Digitization - Dimensions - Width: 3838 pixel / #1 - Digitization - Dimensions - Height: 3710 pixel / #1 - Digitization - Frames/image: 1-40 / #1 - Number grids imaged: 1 / #1 - Number real images: 5950 / #1 - Average exposure time: 10.0 sec. / #1 - Average electron dose: 40.5 e/Å2 / #1 - Details: non-tilted dataset 10 images per "hole" |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -2.5 µm / Nominal defocus min: -1.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)