[English] 日本語
 Yorodumi
Yorodumi- EMDB-12885: Structure of the apo-state of the bacteriophage PhiKZ non-virion ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-12885 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the apo-state of the bacteriophage PhiKZ non-virion RNA polymerase - class including clamp | |||||||||
 Map data Map data | PhiKZ non-virion RNA polymerase - class including clamp | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | RNA polymerase / PhiKZ / non-virion / TRANSCRIPTION | |||||||||
| Function / homology | PHIKZ123 / PHIKZ074 / PHIKZ068 / PHIKZ055 Function and homology information Function and homology information | |||||||||
| Biological species |  Pseudomonas virus phiKZ / Pseudomonas virus phiKZ /  Pseudomonas phage phiKZ (virus) Pseudomonas phage phiKZ (virus) | |||||||||
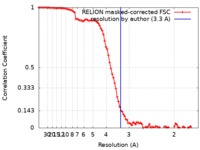
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | de Martin Garrido N / Lai Wan Loong YTE | |||||||||
| Funding support |  United Kingdom, United Kingdom,  Russian Federation, 2 items Russian Federation, 2 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2021 Journal: Nucleic Acids Res / Year: 2021Title: Structure of the bacteriophage PhiKZ non-virion RNA polymerase. Authors: Natàlia deYMartín Garrido / Mariia Orekhova / Yuen Ting Emilie Lai Wan Loong / Anna Litvinova / Kailash Ramlaul / Tatyana Artamonova / Alexei S Melnikov / Pavel Serdobintsev / Christopher ...Authors: Natàlia deYMartín Garrido / Mariia Orekhova / Yuen Ting Emilie Lai Wan Loong / Anna Litvinova / Kailash Ramlaul / Tatyana Artamonova / Alexei S Melnikov / Pavel Serdobintsev / Christopher H S Aylett / Maria Yakunina /   Abstract: Bacteriophage ΦKZ (PhiKZ) is the archetype of a family of massive bacterial viruses. It is considered to have therapeutic potential as its host, Pseudomonas aeruginosa, is an opportunistic, ...Bacteriophage ΦKZ (PhiKZ) is the archetype of a family of massive bacterial viruses. It is considered to have therapeutic potential as its host, Pseudomonas aeruginosa, is an opportunistic, intrinsically antibiotic resistant, pathogen that kills tens of thousands worldwide each year. ΦKZ is an incredibly interesting virus, expressing many systems that the host already possesses. On infection, it forms a 'nucleus', erecting a barrier around its genome to exclude host endonucleases and CRISPR-Cas systems. ΦKZ infection is independent of the host transcriptional apparatus. It expresses two different multi-subunit RNA polymerases (RNAPs): the virion RNAP (vRNAP) is injected with the viral DNA during infection to transcribe early genes, including those encoding the non-virion RNAP (nvRNAP), which transcribes all further genes. ΦKZ nvRNAP is formed by four polypeptides thought to represent homologues of the eubacterial β/β' subunits, and a fifth with unclear homology, but essential for transcription. We have resolved the structure of ΦKZ nvRNAP to better than 3.0 Å, shedding light on its assembly, homology, and the biological role of the fifth subunit: it is an embedded, integral member of the complex, the position, structural homology and biochemical role of which imply that it has evolved from an ancestral homologue to σ-factor. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_12885.map.gz emd_12885.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-12885-v30.xml emd-12885-v30.xml emd-12885.xml emd-12885.xml | 25.9 KB 25.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_12885_fsc.xml emd_12885_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_12885.png emd_12885.png | 131.8 KB | ||
| Filedesc metadata |  emd-12885.cif.gz emd-12885.cif.gz | 8.4 KB | ||
| Others |  emd_12885_half_map_1.map.gz emd_12885_half_map_1.map.gz emd_12885_half_map_2.map.gz emd_12885_half_map_2.map.gz | 49.6 MB 49.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-12885 http://ftp.pdbj.org/pub/emdb/structures/EMD-12885 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12885 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-12885 | HTTPS FTP |
-Related structure data
| Related structure data |  7ogpMC  7ogrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10707 (Title: Bacteriophage PhiKZ non-virion RNA Polymerase / Data size: 769.6 EMPIAR-10707 (Title: Bacteriophage PhiKZ non-virion RNA Polymerase / Data size: 769.6 Data #1: PhiKZ non-virion RNA polymerase particles [micrographs - single frame])  EMPIAR-10709 (Title: Bacteriophage PhiKZ non-virion RNA Polymerase / Data size: 3.3 TB EMPIAR-10709 (Title: Bacteriophage PhiKZ non-virion RNA Polymerase / Data size: 3.3 TBData #1: PhiKZ non-virion RNA polymerase particles [micrographs - multiframe]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_12885.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_12885.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PhiKZ non-virion RNA polymerase - class including clamp | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.85 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: PhiKZ non-virion RNA polymerase - class including clamp - halfmap 1
| File | emd_12885_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PhiKZ non-virion RNA polymerase - class including clamp - halfmap 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
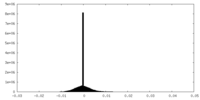
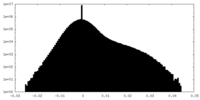
| Density Histograms |
-Half map: PhiKZ non-virion RNA polymerase - class including clamp - halfmap 2
| File | emd_12885_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | PhiKZ non-virion RNA polymerase - class including clamp - halfmap 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
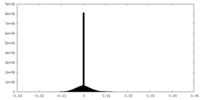
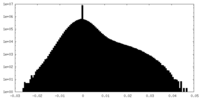
| Density Histograms |
- Sample components
Sample components
-Entire : Bacteriophage PhiKZ non-virion RNA polymerase
| Entire | Name: Bacteriophage PhiKZ non-virion RNA polymerase |
|---|---|
| Components |
|
-Supramolecule #1: Bacteriophage PhiKZ non-virion RNA polymerase
| Supramolecule | Name: Bacteriophage PhiKZ non-virion RNA polymerase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 Details: Expressed in E.coli from sequences obtained from the St. Petersburg lab strain of bacteriophage PhiKZ |
|---|---|
| Source (natural) | Organism:  Pseudomonas virus phiKZ / Strain: St. Petersburg Pseudomonas virus phiKZ / Strain: St. Petersburg |
| Molecular weight | Theoretical: 300 KDa |
-Macromolecule #1: PHIKZ055
| Macromolecule | Name: PHIKZ055 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage phiKZ (virus) Pseudomonas phage phiKZ (virus) |
| Molecular weight | Theoretical: 57.976605 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MGLYAKVVDH NEVHDQFTGK RIYANDYNTS NSDEKEEFDR HFYSHFQDSE AIESSVSCDC RAIEDAHKL GVICDICNTP VVNTSSRPIE PSMWVRTPKH VRSLINPRLI IMLTGYLVTK EFDFLAYLTD TSYRYDVESI G SKETRRKV ...String: MGSSHHHHHH SSGLVPRGSH MGLYAKVVDH NEVHDQFTGK RIYANDYNTS NSDEKEEFDR HFYSHFQDSE AIESSVSCDC RAIEDAHKL GVICDICNTP VVNTSSRPIE PSMWVRTPKH VRSLINPRLI IMLTGYLVTK EFDFLAYLTD TSYRYDVESI G SKETRRKV DRLLHRGFER GLNHFIDNFN EIFQFLLDAN IISNNKSEFA QFVAQNKDKL FPKYLPVPSK LCFVAESTTS GT YLDKPIE AAIDATLTFA SIDASSVPLS PIKAQNRTMR GLRLYGQFYE IYAKSRIAQK PGLARRHMFG ARLNATARAV ITS ISDPHD YDELHIPWGV GCQLLKYHLT NKLKAKFNMT TREAFSFVYE NVLQYNQIIA DLFKELIAEA APYKGMGCTF HRNP TLQRG STQQFFITKV KDDINDNSIS MSVLCLKAPN ADFDGDQLNL TLMPDVYLTK ATERIAPHTW VLSIDEPHEI SGNLE LQGP VVETIINWAH EKYLPPLEEW LKAK UniProtKB: PHIKZ055 |
-Macromolecule #2: PHIKZ068
| Macromolecule | Name: PHIKZ068 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage phiKZ (virus) Pseudomonas phage phiKZ (virus) |
| Molecular weight | Theoretical: 59.41977 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEIIVTGVQG TGFTEVATEH NGKRLTWTTT AYSKIRVQDQ QRVFQEINDY WSGLSAEAQQ HIWNCYVEIR KIMDMAMDPM RIAMSLSYY IKEMYKAMPM NSFRRWLLTI GKLYIPVDIE EVITDDSRYN RPDQTYLKHD YINLASVSLA LRPLVPIWGE F IDQGTSQE ...String: MEIIVTGVQG TGFTEVATEH NGKRLTWTTT AYSKIRVQDQ QRVFQEINDY WSGLSAEAQQ HIWNCYVEIR KIMDMAMDPM RIAMSLSYY IKEMYKAMPM NSFRRWLLTI GKLYIPVDIE EVITDDSRYN RPDQTYLKHD YINLASVSLA LRPLVPIWGE F IDQGTSQE MHKECEVISL ISDCEVNHWP VDEISIDGTP VETAYDKLSA YVKFCVEDEA PTLANLYRGM SSAEVPDILQ AK VMVRRLT ILPLNDATSH SIVSNMFRYV KSNLNPAERS TADRVNDKRP DKGGIDDDDK TSFIESHKTK QRVTPGDIVA YNL DALDVV KLVHKIDDTV PVELIQECLD CVAVTATKDI YPHQILLAQW VMHKAFPARA FSHINKNAVN HLLAAAQSLM WHWG FQQVA VFMQVELYYS GEHAMSIQPR NSTRIQIKYK DVMDELYPHQ RQQRAINGVP VAPVNIAGIA VQSAHASIRS SNWIY HGPD RLFKEAEQVT QNKVLVVPAT IKSVITELVI HLGKLNQ UniProtKB: PHIKZ068 |
-Macromolecule #3: DNA-directed RNA polymerase
| Macromolecule | Name: DNA-directed RNA polymerase / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed RNA polymerase |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage phiKZ (virus) Pseudomonas phage phiKZ (virus) |
| Molecular weight | Theoretical: 78.780453 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSQLGRREID LTLLGHTGLD PWYGTTSSAR GAMFVTHIGQ APEVNGNESR YFLTGAELEY AKYTHDVRFP EDCRVLHVLR KYPTGIGKD SIRSNPVTTI IYENYFDKYK TIGVLHVPEY MSHHQDFGYE LVKNREVWET IAPNEMFSKD TVIAQSGAVK K DGTLGMGV ...String: MSQLGRREID LTLLGHTGLD PWYGTTSSAR GAMFVTHIGQ APEVNGNESR YFLTGAELEY AKYTHDVRFP EDCRVLHVLR KYPTGIGKD SIRSNPVTTI IYENYFDKYK TIGVLHVPEY MSHHQDFGYE LVKNREVWET IAPNEMFSKD TVIAQSGAVK K DGTLGMGV NANVVFLSAA GTIEDGFVAN KNFLKRMMPT SYSTAVANAG RKAFFLNMYG DDKIYKPFPD IGDVIRPDGV IF AIRDHDD DLAPAEMTPR ALRTLDRTFD RAVIGTPGAK VIDIDIWRDE RVNPSPTPTG MDAQLVKYHT HLSSYYRELL KIY RGLLAR RKDDLHITEE FERLIVTAQM FLPQPDNVRK LSRFYRLDPL DEWRVEVTYK AQKMPAGAFK MTDFHGGKGV ICKV MEDED MPIDENGNRA DLIIFGGSTM RRSNYGRIYE HGFGAAARDL AQRLRVEAGL DRHAKPTQQQ LNSVMGNTQW VDYAF KELL GFYEIIAPTM HSKMMEHPNP AEHVKTVLMD GFPYIYAPVD DPVDLMAAVN KLINSDKYRP HYGKVSYRDQ AGKWVT TKD NVLMGPLYMM LLEKIGEDWS AAASVKTQPF GLPSKLNNAD RASTPGRETA IRSFGESETR SYNCTVGPGP TAEILDQ TN NPLAHAAVIE SWLTAEKPSS VPVAVDREKI PFGGSRPVAM FDHLLECSGI ALEYAPDH |
-Macromolecule #4: PHIKZ074
| Macromolecule | Name: PHIKZ074 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage phiKZ (virus) Pseudomonas phage phiKZ (virus) |
| Molecular weight | Theoretical: 77.513461 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNLNRYKARD LLNLSYDDLW SLPSEWHLIE FDDGKTVVSV DRITKLSVLC WYPLKHYKDC PIPSDHHIDF NRILTDNPKD YLNVEGGRV TSKAMVKHLN KAIWNIYDWS GETVDPEVLS KLAIEGKNWL YNQTTVKLSE YLATLSMFDI AEVYNHPKVR E ANHNIEPT ...String: MNLNRYKARD LLNLSYDDLW SLPSEWHLIE FDDGKTVVSV DRITKLSVLC WYPLKHYKDC PIPSDHHIDF NRILTDNPKD YLNVEGGRV TSKAMVKHLN KAIWNIYDWS GETVDPEVLS KLAIEGKNWL YNQTTVKLSE YLATLSMFDI AEVYNHPKVR E ANHNIEPT TYGIEKISYG KVKEVFNDPT QFIGNSIIEG LRSGTQKTEQ LLQAFAWRGF PTDINSDIFK YPVTTGYIDG IW NLYENMI ESRSGTKALL YNKELLRVTE YFNRKSQLIA QYVQRLHPGD CKTTILAEYP VTKLTLKAFK GKYYQKEDGK LDW IRGNET HLIGTKQKFR SVFGCNHPDS QGICMTCYGR LGINIPKGTN IGQVAAVSMG DKITSAVLST KHTDASSAVE QYKL GKIES NYLRTGEIPE TLYLKKELTQ KDYRLVIARS EAENLADILM IDDLTAYPAT SATELTSLAL VYDDEVNGEC GDVLT VSLY NRRASLSIEM LKHIKMVRWE LDQRDNIVIS LRGFDFNLPF LTLPNKHVNM YEVMKRFQSF LHSGSDSAEA GKLSTE KVG YTSKTYLKNY KSPIEALPVF ATMANEKISL NISHCEILIY AMMIRSAQYR DYRLPKPGIN GQFEKYNRLM QCRSLGG AM AFEKQHEPLN NPGSFLNKMR NDHPYDLLVK GGKLR UniProtKB: PHIKZ074 |
-Macromolecule #5: PHIKZ123
| Macromolecule | Name: PHIKZ123 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas phage phiKZ (virus) Pseudomonas phage phiKZ (virus) |
| Molecular weight | Theoretical: 62.95909 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MPDPFLIEKI RENTPCMNPT LANGITVEHT MTRDPNTGVN MTRRYIDSLF DISSVLFPDG FKYEGNRACT PLKHFEEITR EYNAKRIAN IAPTDMYMID LMFSYKGEML YPRPMLLPAF KRGNMVTING AKYIGSPVLT DVGFSVLNDS IFIPFRRTKL T FKQTDHHY ...String: MPDPFLIEKI RENTPCMNPT LANGITVEHT MTRDPNTGVN MTRRYIDSLF DISSVLFPDG FKYEGNRACT PLKHFEEITR EYNAKRIAN IAPTDMYMID LMFSYKGEML YPRPMLLPAF KRGNMVTING AKYIGSPVLT DVGFSVLNDS IFIPFRRTKL T FKQTDHHY MCNGQRKIMY VIWSQIHNEM AKRTKRDLGN RPHIESCLAH YFFCQFGVTQ TFKQWANVDV KCGLLSDFPE EE YPREKWN IYSSATLKGK HPTGEMVLVI PRHQESIFAT RLIAGFWYVV DAFPMRFTRP EYVDSTNLWR VILGHMVFGD FEH QGKVEE NIDSHLHSFC NSLDEMTIEE LKTVGVNVST IWELLYEIMT SLAHHLYATD IDETSMYGKR LTVLHYLMSE FNYA VSMFG YMFQSRRDRE WTVQELNEGL KRSFKLQTAI KRLTVDHGEL DTMSNPNSSM LIKGTSILVT QDRAKTAKAH NKSLI NDSS RIIHASIAEV GQYKNQPKNN PDGRGRLNMY TKVGPTGLVE RREEVREIID NAQLMFRAK UniProtKB: PHIKZ123 |
-Macromolecule #6: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.3 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 15 mM Tris-Cl pH 8.0, 150 mM NaCl, 0.5 mM EDTA, 2 mM MgCl2, 1 mM DTT |
| Grid | Model: Quantifoil R2/1 / Support film - Material: GRAPHENE OXIDE / Support film - topology: CONTINUOUS / Support film - Film thickness: 1 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 278 K / Instrument: FEI VITROBOT MARK IV / Details: 1-2 s blot. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 100.0 K / Max: 100.0 K |
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 70 eV |
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Digitization - Dimensions - Width: 5096 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 2 / Number real images: 10582 / Average electron dose: 40.0 e/Å2 / Details: TIFF movie mode |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.25 µm / Nominal defocus min: 0.75 µm / Nominal magnification: 48000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Source name: PDB / Chain - Initial model type: experimental model |
|---|---|
| Details | Core conserved regions initially identified from comparison with PDB 6EDT - remainder built de novo |
| Refinement | Protocol: OTHER |
| Output model |  PDB-7ogp: |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)