[English] 日本語
 Yorodumi
Yorodumi- EMDB-10616: FAK structure with AMP-PNP from single particle analysis of 2D cr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-10616 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | FAK structure with AMP-PNP from single particle analysis of 2D crystals | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Kinase / Focal Adhesion / Membrane / CELL ADHESION | |||||||||
| Function / homology |  Function and homology information Function and homology informationApoptotic cleavage of cellular proteins / NCAM signaling for neurite out-growth / RAF/MAP kinase cascade / Turbulent (oscillatory, disturbed) flow shear stress activates signaling by PIEZO1 and integrins in endothelial cells / RHO GTPases Activate WASPs and WAVEs / Regulation of actin dynamics for phagocytic cup formation / negative regulation of protein autophosphorylation / radial glia-guided pyramidal neuron migration / calcium-dependent cysteine-type endopeptidase activity / positive regulation of substrate-dependent cell migration, cell attachment to substrate ...Apoptotic cleavage of cellular proteins / NCAM signaling for neurite out-growth / RAF/MAP kinase cascade / Turbulent (oscillatory, disturbed) flow shear stress activates signaling by PIEZO1 and integrins in endothelial cells / RHO GTPases Activate WASPs and WAVEs / Regulation of actin dynamics for phagocytic cup formation / negative regulation of protein autophosphorylation / radial glia-guided pyramidal neuron migration / calcium-dependent cysteine-type endopeptidase activity / positive regulation of substrate-dependent cell migration, cell attachment to substrate / Integrin signaling / GRB2:SOS provides linkage to MAPK signaling for Integrins / : / MET activates PTK2 signaling / Extra-nuclear estrogen signaling / EPHB-mediated forward signaling / p130Cas linkage to MAPK signaling for integrins / VEGFA-VEGFR2 Pathway / signal complex assembly / response to pH / angiogenesis involved in wound healing / wound healing, spreading of cells / positive regulation of protein tyrosine kinase activity / negative regulation of anoikis / negative regulation of cell-substrate adhesion / positive regulation of focal adhesion assembly / positive regulation of protein binding / regulation of cell adhesion / response to muscle stretch / molecular function activator activity / actin filament organization / non-membrane spanning protein tyrosine kinase activity / non-specific protein-tyrosine kinase / sarcolemma / integrin binding / epidermal growth factor receptor signaling pathway / protein autophosphorylation / protease binding / protein tyrosine kinase activity / cell cortex / positive regulation of cell migration / ciliary basal body / focal adhesion / positive regulation of cell population proliferation / centrosome / perinuclear region of cytoplasm / ATP binding / identical protein binding / nucleus / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
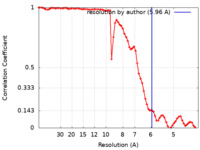
| Method | single particle reconstruction / cryo EM / Resolution: 5.96 Å | |||||||||
 Authors Authors | Acebron I / Righetto R | |||||||||
| Funding support |  Spain, Spain,  Switzerland, 2 items Switzerland, 2 items
| |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2020 Journal: EMBO J / Year: 2020Title: Structural basis of Focal Adhesion Kinase activation on lipid membranes. Authors: Iván Acebrón / Ricardo D Righetto / Christina Schoenherr / Svenja de Buhr / Pilar Redondo / Jayne Culley / Carlos F Rodríguez / Csaba Daday / Nikhil Biyani / Oscar Llorca / Adam Byron / ...Authors: Iván Acebrón / Ricardo D Righetto / Christina Schoenherr / Svenja de Buhr / Pilar Redondo / Jayne Culley / Carlos F Rodríguez / Csaba Daday / Nikhil Biyani / Oscar Llorca / Adam Byron / Mohamed Chami / Frauke Gräter / Jasminka Boskovic / Margaret C Frame / Henning Stahlberg / Daniel Lietha /     Abstract: Focal adhesion kinase (FAK) is a key component of the membrane proximal signaling layer in focal adhesion complexes, regulating important cellular processes, including cell migration, proliferation, ...Focal adhesion kinase (FAK) is a key component of the membrane proximal signaling layer in focal adhesion complexes, regulating important cellular processes, including cell migration, proliferation, and survival. In the cytosol, FAK adopts an autoinhibited state but is activated upon recruitment into focal adhesions, yet how this occurs or what induces structural changes is unknown. Here, we employ cryo-electron microscopy to reveal how FAK associates with lipid membranes and how membrane interactions unlock FAK autoinhibition to promote activation. Intriguingly, initial binding of FAK to the membrane causes steric clashes that release the kinase domain from autoinhibition, allowing it to undergo a large conformational change and interact itself with the membrane in an orientation that places the active site toward the membrane. In this conformation, the autophosphorylation site is exposed and multiple interfaces align to promote FAK oligomerization on the membrane. We show that interfaces responsible for initial dimerization and membrane attachment are essential for FAK autophosphorylation and resulting cellular activity including cancer cell invasion, while stable FAK oligomerization appears to be needed for optimal cancer cell proliferation in an anchorage-independent manner. Together, our data provide structural details of a key membrane bound state of FAK that is primed for efficient autophosphorylation and activation, hence revealing the critical event in integrin mediated FAK activation and signaling at focal adhesions. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_10616.map.gz emd_10616.map.gz | 27.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-10616-v30.xml emd-10616-v30.xml emd-10616.xml emd-10616.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_10616_fsc.xml emd_10616_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_10616.png emd_10616.png | 223.1 KB | ||
| Masks |  emd_10616_msk_1.map emd_10616_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-10616.cif.gz emd-10616.cif.gz | 6.2 KB | ||
| Others |  emd_10616_half_map_1.map.gz emd_10616_half_map_1.map.gz emd_10616_half_map_2.map.gz emd_10616_half_map_2.map.gz | 11.7 MB 11.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-10616 http://ftp.pdbj.org/pub/emdb/structures/EMD-10616 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10616 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-10616 | HTTPS FTP |
-Validation report
| Summary document |  emd_10616_validation.pdf.gz emd_10616_validation.pdf.gz | 916.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_10616_full_validation.pdf.gz emd_10616_full_validation.pdf.gz | 916.4 KB | Display | |
| Data in XML |  emd_10616_validation.xml.gz emd_10616_validation.xml.gz | 12.7 KB | Display | |
| Data in CIF |  emd_10616_validation.cif.gz emd_10616_validation.cif.gz | 17.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10616 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10616 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10616 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-10616 | HTTPS FTP |
-Related structure data
| Related structure data |  6ty4MC  6ty3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10347 (Title: FAK structure from single particle analysis of 2D crystals EMPIAR-10347 (Title: FAK structure from single particle analysis of 2D crystalsData size: 1.8 TB Data #1: Dose-weighted aligned movie averages of the FAK AMP-PNP dataset [micrographs - single frame] Data #2: Unaligned movies of the FAK AMP-PNP dataset [micrographs - multiframe] Data #3: Stack of particles extracted from the FAK AMP-PNP dataset [picked particles - single frame - unprocessed] Data #4: Dose-weighted aligned movie averages of the FAK APO Polara dataset [micrographs - single frame] Data #5: Dose-weighted aligned movie averages of the FAK APO Titan dataset [micrographs - single frame] Data #6: Unaligned movies of the FAK APO Titan dataset [micrographs - multiframe] Data #7: Stack of particles extracted from the FAK APO Titan and Polara datasets [picked particles - single frame - unprocessed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_10616.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_10616.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.116 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_10616_msk_1.map emd_10616_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_10616_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_10616_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Focal adhesion kinase
| Entire | Name: Focal adhesion kinase |
|---|---|
| Components |
|
-Supramolecule #1: Focal adhesion kinase
| Supramolecule | Name: Focal adhesion kinase / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 150 KDa |
-Macromolecule #1: Focal adhesion kinase 1
| Macromolecule | Name: Focal adhesion kinase 1 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: non-specific protein-tyrosine kinase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 75.476445 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: GPGAMERVLK VFHYFENSSE PTTWASIIRH GDATDVRGII QKIVDCHKVK NVACYGLRLS HLQSEEVHWL HLDMGVSNVR EKFELAHPP EEWKYELRIR YLPKGFLNQF TEDKPTLNFF YQQVKNDYML EIADQVDQEI ALKLGCLEIR RSYGEMRGNA L EKKSNYEV ...String: GPGAMERVLK VFHYFENSSE PTTWASIIRH GDATDVRGII QKIVDCHKVK NVACYGLRLS HLQSEEVHWL HLDMGVSNVR EKFELAHPP EEWKYELRIR YLPKGFLNQF TEDKPTLNFF YQQVKNDYML EIADQVDQEI ALKLGCLEIR RSYGEMRGNA L EKKSNYEV LEKDVGLRRF FPKSLLDSVK AKTLRKLIQQ TFRQFANLNR EESILKFFEI LSPVYRFDKE CFKCALGSSW II SVELAIG PEEGISYLTD KGANPTHLAD FNQVQTIQYS NSEDKDRKGM LQLKIAGAPE PLTVTAPSLT IAENMADLID GYC RLVNGA TQSFIIRPQK EGERALPSIP KLANNEKQGV RSHTVSVSET DDYAEIIDEE DTYTMPSTRD YEIQRERIEL GRCI GEGQF GDVHQGIYMS PENPAMAVAI KTCKNCTSDS VREKFLQEAL TMRQFDHPHI VKLIGVITEN PVWIIMELCT LGELR SFLQ VRKFSLDLAS LILYAYQLST ALAYLESKRF VHRDIAARNV LVSATDCVKL GDFGLSRYME DSTYYKASKG KLPIKW MAP ESINFRRFTS ASDVWMFGVC MWEILMHGVK PFQGVKNNDV IGRIENGERL PMPPNCPPTL YSLMTKCWAY DPSRRPR FT ELKAQLSTIL EEEKLQ UniProtKB: Focal adhesion kinase 1 |
-Macromolecule #2: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 2 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #3: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 3 / Number of copies: 2 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | 2D array |
- Sample preparation
Sample preparation
| Buffer | pH: 5.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT | ||||||||
| Output model |  PDB-6ty4: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)