+ Open data
Open data
- Basic information
Basic information
| Entry | 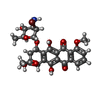 Database: PDB chemical components / ID: DM8 Database: PDB chemical components / ID: DM8 |
|---|---|
| Name | Name: Synonyms: WP401; DAUNOMYCIN DERIVATIVE; DAUNORUBICIN DERIVATIVE |
-Chemical information
| Composition |  | ||||||
|---|---|---|---|---|---|---|---|
| Others | Type: NON-POLYMER / PDB classification: HETAD / Three letter code: DM8 / Model coordinates PDB-ID: 288D | ||||||
| History |
| ||||||
 External links External links |  UniChem / UniChem /  ChemSpider / ChemSpider /  Nikkaji / Nikkaji /  PubChem / PubChem /  SureChEMBL / SureChEMBL /  ZINC / ZINC /  Wikipedia search / Wikipedia search /  Google search Google search |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
-Details
-SMILES
| ACDLabs 10.04 | | CACTVS 3.341 | OpenEye OEToolkits 1.5.0 | |
|---|
-SMILES CANONICAL
| CACTVS 3.341 | | OpenEye OEToolkits 1.5.0 | |
|---|
-InChI
| InChI 1.03 |
|---|
-InChIKey
| InChI 1.03 |
|---|
-SYSTEMATIC NAME
| ACDLabs 10.04 | (| OpenEye OEToolkits 1.5.0 | ( | |
|---|
-PDB entries
Showing all 6 items
PDB-276d: 
SUBSTITUTIONS AT C2' OF DAUNOSAMINE IN THE ANTICANCER DAUNORUBICIN ALTER ITS DNA-BINDING SEQUENCE SPECIFICITY
PDB-277d: 
SUBSTITUTIONS AT C2' OF DAUNOSAMINE IN THE ANTICANCER DAUNORUBICIN ALTER ITS DNA-BINDING SEQUENCE SPECIFICITY
PDB-278d: 
SUBSTITUTIONS AT C2' OF DAUNOSAMINE IN THE ANTICANCER DAUNORUBICIN ALTER ITS DNA-BINDING SEQUENCE SPECIFICITY
PDB-288d: 
SUBSTITUTIONS AT C2' OF DAUNOSAMINE IN THE ANTICANCER DAUNORUBICIN ALTER ITS DNA-BINDING SEQUENCE SPECIFICITY
PDB-380d: 
BINDING OF THE MODIFIED DAUNORUBICIN WP401 ADJACENT TO A T-G BASE PAIR INDUCES THE REVERSE WATSON-CRICK CONFORMATION: CRYSTAL STRUCTURES OF THE WP401-TGGCCG AND WP401-CGG[BR5C]CG COMPLEXES
PDB-381d: 
BINDING OF THE MODIFIED DAUNORUBICIN WP401 ADJACENT TO A T-G BASE PAIR INDUCES THE REVERSE WATSON-CRICK CONFORMATION: CRYSTAL STRUCTURES OF THE WP401-TGGCCG AND WP401-CGG[BR5C]CG COMPLEXES
 Movie
Movie Controller
Controller



