+ Open data
Open data
- Basic information
Basic information
| Entry | 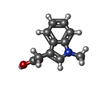 Database: PDB chemical components / ID: 3VY Database: PDB chemical components / ID: 3VY |
|---|---|
| Name | Name: ( |
-Chemical information
| Composition |  | ||||
|---|---|---|---|---|---|
| Others | Type: NON-POLYMER / PDB classification: HETAIN / Three letter code: 3VY / Ideal coordinates details: Corina / Model coordinates PDB-ID: 4WXP | ||||
| History |
| ||||
 External links External links |  UniChem / UniChem /  ChemSpider / ChemSpider /  Wikipedia search / Wikipedia search /  Google search Google search |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
-Details
-SMILES
| ACDLabs 12.01 | | CACTVS 3.385 | OpenEye OEToolkits 1.9.2 | |
|---|
-SMILES CANONICAL
| CACTVS 3.385 | | OpenEye OEToolkits 1.9.2 | |
|---|
-InChI
| InChI 1.03 |
|---|
-InChIKey
| InChI 1.03 |
|---|
-SYSTEMATIC NAME
| ACDLabs 12.01 | (| OpenEye OEToolkits 1.9.2 | |
|---|
-PDB entries
Showing all 2 items

PDB-4wxp: 
X-ray crystal structure of NS3 Helicase from HCV with a bound fragment inhibitor at 2.08 A resolution

PDB-5fpt: 
Structure of hepatitis C virus (HCV) full-length NS3 complex with small-molecule ligand 2-(1-methyl-1H-indol-3-yl)acetic acid (AT3437) in an alternate binding site.
 Movie
Movie Controller
Controller



