+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9673 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Mud Crab Dicistrovirus | |||||||||
 Map data Map data | ||||||||||
 Sample Sample | Mud crab dicistrovirus != Mud crab virus Mud crab dicistrovirus
| |||||||||
 Keywords Keywords | Dicistrovirus / VIRUS | |||||||||
| Function / homology | Dicistrovirus, capsid-polyprotein, C-terminal / CRPV capsid protein like / virion component / Picornavirus/Calicivirus coat protein / Viral coat protein subunit / Structural polyprotein Function and homology information Function and homology information | |||||||||
| Biological species |  Mud crab virus Mud crab virus | |||||||||
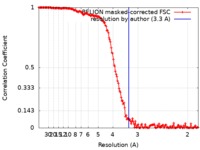
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Zhang Q / Gao Y | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: J Virol / Year: 2019 Journal: J Virol / Year: 2019Title: Cryo-electron Microscopy Structures of Novel Viruses from Mud Crab with Multiple Infections. Authors: Yuanzhu Gao / Shanshan Liu / Jiamiao Huang / Qianqian Wang / Kunpeng Li / Jian He / Jianguo He / Shaoping Weng / Qinfen Zhang /  Abstract: Viruses associated with sleeping disease (SD) in crabs cause great economic losses to aquaculture, and no effective measures are available for their prevention. In this study, to help develop novel ...Viruses associated with sleeping disease (SD) in crabs cause great economic losses to aquaculture, and no effective measures are available for their prevention. In this study, to help develop novel antiviral strategies, single-particle cryo-electron microscopy was applied to investigate viruses associated with SD. The results not only revealed the structure of mud crab dicistrovirus (MCDV) but also identified a novel mud crab tombus-like virus (MCTV) not previously detected using molecular biology methods. The structure of MCDV at a 3.5-Å resolution reveals three major capsid proteins (VP1 to VP3) organized into a pseudo-T=3 icosahedral capsid, and affirms the existence of VP4. Unusually, MCDV VP3 contains a long C-terminal region and forms a novel protrusion that has not been observed in other dicistrovirus. Our results also reveal that MCDV can release its genome via conformation changes of the protrusions when viral mixtures are heated. The structure of MCTV at a 3.3-Å resolution reveals a T= 3 icosahedral capsid with common features of both tombusviruses and nodaviruses. Furthermore, MCTV has a novel hydrophobic tunnel beneath the 5-fold vertex and 30 dimeric protrusions composed of the P-domains of the capsid protein at the 2-fold axes that are exposed on the virion surface. The structural features of MCTV are consistent with a novel type of virus. Pathogen identification is vital for unknown infectious outbreaks, especially for dual or multiple infections. Sleeping disease (SD) in crabs causes great economic losses to aquaculture worldwide. Here we report the discovery and identification of a novel virus in mud crabs with multiple infections that was not previously detected by molecular, immune, or traditional electron microscopy (EM) methods. High-resolution structures of pathogenic viruses are essential for a molecular understanding and developing new disease prevention methods. The three-dimensional (3D) structure of the mud crab tombus-like virus (MCTV) and mud crab dicistrovirus (MCDV) determined in this study could assist the development of antiviral inhibitors. The identification of a novel virus in multiple infections previously missed using other methods demonstrates the usefulness of this strategy for investigating multiple infectious outbreaks, even in humans and other animals. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9673.map.gz emd_9673.map.gz | 397.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9673-v30.xml emd-9673-v30.xml emd-9673.xml emd-9673.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9673_fsc.xml emd_9673_fsc.xml | 15.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_9673.png emd_9673.png | 334.8 KB | ||
| Filedesc metadata |  emd-9673.cif.gz emd-9673.cif.gz | 6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9673 http://ftp.pdbj.org/pub/emdb/structures/EMD-9673 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9673 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9673 | HTTPS FTP |
-Validation report
| Summary document |  emd_9673_validation.pdf.gz emd_9673_validation.pdf.gz | 751.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9673_full_validation.pdf.gz emd_9673_full_validation.pdf.gz | 751 KB | Display | |
| Data in XML |  emd_9673_validation.xml.gz emd_9673_validation.xml.gz | 15.6 KB | Display | |
| Data in CIF |  emd_9673_validation.cif.gz emd_9673_validation.cif.gz | 21.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9673 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9673 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9673 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9673 | HTTPS FTP |
-Related structure data
| Related structure data |  6iicMC  9754C  9755C  9756C  6izlC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9673.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9673.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.933 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Mud crab dicistrovirus
| Entire | Name:  Mud crab dicistrovirus Mud crab dicistrovirus |
|---|---|
| Components |
|
-Supramolecule #1: Mud crab virus
| Supramolecule | Name: Mud crab virus / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 932662 / Sci species name: Mud crab virus / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Scylla paramamosain (green mud crab) Scylla paramamosain (green mud crab) |
-Macromolecule #1: VP1 of Mud crab dicistrovirus
| Macromolecule | Name: VP1 of Mud crab dicistrovirus / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mud crab virus Mud crab virus |
| Molecular weight | Theoretical: 20.691158 KDa |
| Sequence | String: GGGMDYAKSD SSSLVTTMGE QFRSLRMLTR RSSPTDVLTG TSVTLPGITI GTDSSLRQSV LNIISYMYRF TKGSISYKII PKIKGDLYI TTSSADNIEL NSNAYSFDVN RALHYQNTAL NPVVQVSLPY YCPSENLVID STSFPNLSNL VLTNLERSSN T YTVLVSAG DDHTFSQLAG CPAFTIGPSR SAA UniProtKB: Structural polyprotein |
-Macromolecule #2: VP2 of Mud crab dicistrovirus
| Macromolecule | Name: VP2 of Mud crab dicistrovirus / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mud crab virus Mud crab virus |
| Molecular weight | Theoretical: 28.074658 KDa |
| Sequence | String: MDSSVTNTGG LMPSATISNS EGATMLLNDI PDPTQNVFLS RNVTDNLFEV QDQNLIESLS REVLLGTGTW QSGQAEISTT LTEQQLITN YEQPSIRQIS LPDDIVKGSS FIASKLANIA YMRCDYELYL RVQGSPFLQG LLLLWNKMNA DQTSKIRSSI T EHLRSITS ...String: MDSSVTNTGG LMPSATISNS EGATMLLNDI PDPTQNVFLS RNVTDNLFEV QDQNLIESLS REVLLGTGTW QSGQAEISTT LTEQQLITN YEQPSIRQIS LPDDIVKGSS FIASKLANIA YMRCDYELYL RVQGSPFLQG LLLLWNKMNA DQTSKIRSSI T EHLRSITS FPGVTLNMQS DSRSVKLVIP YTSEFQVFNP RNENKLNSVR LSILSALRGP STSEKATYSI MGRMTNIKLY GH APSIVSL SYPQTE UniProtKB: Structural polyprotein |
-Macromolecule #3: VP3 of Mud crab dicistrovirus
| Macromolecule | Name: VP3 of Mud crab dicistrovirus / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mud crab virus Mud crab virus |
| Molecular weight | Theoretical: 48.24968 KDa |
| Sequence | String: SKDRDLSKVS PYENIPAKGF THGVGFDYGV PLSLFPDNAI DPTIANPESI DEMSIQYLAS RPYMLDRYTI KGGNTPSPSG TVVADIPIS PVNYSLYGSI IRDYRTIFGA PVSLAVAMAS WWRAKIHLNL QFAKTQYHQC RLLVQYLPYG SDVQSLENVL S QIIDISHV ...String: SKDRDLSKVS PYENIPAKGF THGVGFDYGV PLSLFPDNAI DPTIANPESI DEMSIQYLAS RPYMLDRYTI KGGNTPSPSG TVVADIPIS PVNYSLYGSI IRDYRTIFGA PVSLAVAMAS WWRAKIHLNL QFAKTQYHQC RLLVQYLPYG SDVQSLENVL S QIIDISHV DESGIDLCFP SIFTNKWMRS YDPATEGYTA GCAPGRILIS VLNPLISAST VNDDIVMMPW LTWENLELAE PG SLAKAAI GFDYPADAVD EKWTSRELPV TGSSFNLFRD TTIVLGASTN ISNLVLTNDD TGGDYQIVST TPTGSYVSAV TCP QGTYTI THDGVGATII SNFPILGAGE GPSFQISALR HGDKVTITED PTKINVSGVS FLTGTNSWKA SLKDSSGTLL GRLE YDGTS FSSDSPASLI PGKYNVELDP ADNSAVVTIV ANNSFGTASL DTH UniProtKB: Structural polyprotein |
-Macromolecule #4: VP4 of Mud crab dicistrovirus
| Macromolecule | Name: VP4 of Mud crab dicistrovirus / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mud crab virus Mud crab virus |
| Molecular weight | Theoretical: 5.792425 KDa |
| Sequence | String: GGDDATASQR GIVTQVADTV SSISNVVDGL GVPLLSSISK PIGWVSNVVS NVASIFGF UniProtKB: Structural polyprotein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Pressure: 101.325 kPa |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 80.0 K / Max: 100.0 K |
| Image recording | Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Digitization - Dimensions - Width: 4096 pixel / Digitization - Dimensions - Height: 4096 pixel / Number grids imaged: 3 / Average exposure time: 1.0 sec. / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 3.0 µm / Calibrated defocus min: 1.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 96000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-6iic: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)