+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-0130 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the Macrobrachium rosenbergii Nodavirus | |||||||||
 Map data Map data | CryoEM reconstruction of Macrobrachium rosenbergii nodavirus virion | |||||||||
 Sample Sample |
| |||||||||
| Biological species | Macrobrachium rosen (others) | |||||||||
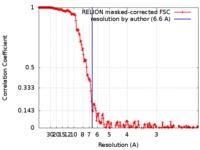
| Method | single particle reconstruction / cryo EM / Resolution: 6.6 Å | |||||||||
 Authors Authors | Ho KH / Gabrielsen M | |||||||||
 Citation Citation |  Journal: PLoS Biol / Year: 2018 Journal: PLoS Biol / Year: 2018Title: Structure of the Macrobrachium rosenbergii nodavirus: A new genus within the Nodaviridae? Authors: Kok Lian Ho / Mads Gabrielsen / Poay Ling Beh / Chare Li Kueh / Qiu Xian Thong / James Streetley / Wen Siang Tan / David Bhella /   Abstract: Macrobrachium rosenbergii nodavirus (MrNV) is a pathogen of freshwater prawns that poses a threat to food security and causes significant economic losses in the aquaculture industries of many ...Macrobrachium rosenbergii nodavirus (MrNV) is a pathogen of freshwater prawns that poses a threat to food security and causes significant economic losses in the aquaculture industries of many developing nations. A detailed understanding of the MrNV virion structure will inform the development of strategies to control outbreaks. The MrNV capsid has also been engineered to display heterologous antigens, and thus knowledge of its atomic resolution structure will benefit efforts to develop tools based on this platform. Here, we present an atomic-resolution model of the MrNV capsid protein (CP), calculated by cryogenic electron microscopy (cryoEM) of MrNV virus-like particles (VLPs) produced in insect cells, and three-dimensional (3D) image reconstruction at 3.3 Å resolution. CryoEM of MrNV virions purified from infected freshwater prawn post-larvae yielded a 6.6 Å resolution structure, confirming the biological relevance of the VLP structure. Our data revealed that unlike other known nodavirus structures, which have been shown to assemble capsids having trimeric spikes, MrNV assembles a T = 3 capsid with dimeric spikes. We also found a number of surprising similarities between the MrNV capsid structure and that of the Tombusviridae: 1) an extensive network of N-terminal arms (NTAs) lines the capsid interior, forming long-range interactions to lace together asymmetric units; 2) the capsid shell is stabilised by 3 pairs of Ca2+ ions in each asymmetric unit; 3) the protruding spike domain exhibits a very similar fold to that seen in the spikes of the tombusviruses. These structural similarities raise questions concerning the taxonomic classification of MrNV. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_0130.map.gz emd_0130.map.gz | 473.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-0130-v30.xml emd-0130-v30.xml emd-0130.xml emd-0130.xml | 12.1 KB 12.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_0130_fsc.xml emd_0130_fsc.xml | 18.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_0130.png emd_0130.png | 246.5 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-0130 http://ftp.pdbj.org/pub/emdb/structures/EMD-0130 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0130 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-0130 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_0130.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_0130.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM reconstruction of Macrobrachium rosenbergii nodavirus virion | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.11 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Macrobrachium rosen
| Entire | Name: Macrobrachium rosen (others) |
|---|---|
| Components |
|
-Supramolecule #1: Macrobrachium rosen
| Supramolecule | Name: Macrobrachium rosen / type: virus / ID: 1 / Parent: 0 Details: Virions were purified from Macrobrachium rosenbergii infected post-larvae NCBI-ID: 32644 / Sci species name: Macrobrachium rosen / Virus type: VIRION / Virus isolate: OTHER / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Macrobrachium rosenbergii (giant freshwater prawn) Macrobrachium rosenbergii (giant freshwater prawn) |
| Virus shell | Shell ID: 1 / Name: Capsid / Diameter: 400.0 Å / T number (triangulation number): 3 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL |
|---|---|
| Buffer | pH: 7.4 / Details: 25mM HEPES, 150 mM NaCl; pH 7.4 |
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Atmosphere: AIR |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV Details: Virions were loaded onto Quantifoil R2/2 grids which had a continuous carbon film added. Virions were adsorbed to the carbon for 1 minute prior to blotting (4s).. |
| Details | Purified virions |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2200FS |
|---|---|
| Specialist optics | Energy filter - Name: In-column Omega Filter / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: DIRECT ELECTRON DE-20 (5k x 3k) / Detector mode: INTEGRATING / Digitization - Frames/image: 2-40 / Number real images: 263 / Average exposure time: 2.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: GATAN 626 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)