[English] 日本語
 Yorodumi
Yorodumi- EMDB-9614: Negative stain EM reconstruction of Mycobacterium Mfd Oligomer (D... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9614 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Negative stain EM reconstruction of Mycobacterium Mfd Oligomer (Dodecamer) | |||||||||
 Map data Map data | 1.5 sigma | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) | |||||||||
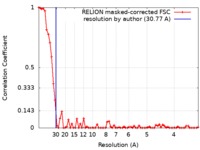
| Method | single particle reconstruction / negative staining / Resolution: 30.77 Å | |||||||||
 Authors Authors | Vinayak B / Putta S / Rao DN / Nagaraja V / Natesh R | |||||||||
| Funding support |  India, 1 items India, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd Authors: Putta S / Prabha S / Bhat V / Fox GC / Walsh MA / Rao DN / Nagaraja V / Natesh R | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9614.map.gz emd_9614.map.gz | 19.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9614-v30.xml emd-9614-v30.xml emd-9614.xml emd-9614.xml | 13.5 KB 13.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9614_fsc.xml emd_9614_fsc.xml | 6.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_9614.png emd_9614.png | 42.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9614 http://ftp.pdbj.org/pub/emdb/structures/EMD-9614 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9614 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9614 | HTTPS FTP |
-Validation report
| Summary document |  emd_9614_validation.pdf.gz emd_9614_validation.pdf.gz | 78.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9614_full_validation.pdf.gz emd_9614_full_validation.pdf.gz | 77.7 KB | Display | |
| Data in XML |  emd_9614_validation.xml.gz emd_9614_validation.xml.gz | 495 B | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9614 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9614 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9614 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9614 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9614.map.gz / Format: CCP4 / Size: 26.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9614.map.gz / Format: CCP4 / Size: 26.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 1.5 sigma | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Mycobacterium tuberculosis Mfd Oligomer (Dodecamer)
| Entire | Name: Mycobacterium tuberculosis Mfd Oligomer (Dodecamer) |
|---|---|
| Components |
|
-Supramolecule #1: Mycobacterium tuberculosis Mfd Oligomer (Dodecamer)
| Supramolecule | Name: Mycobacterium tuberculosis Mfd Oligomer (Dodecamer) / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: Oligomeric MtbMfd fraction from Size Exclusion Chromatography. |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) / Strain: H37Rv Mycobacterium tuberculosis H37Rv (bacteria) / Strain: H37Rv |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 1.596 MDa |
-Macromolecule #1: Mycobactetrium tuberculosis Mfd Oligomer (Dodecamer)
| Macromolecule | Name: Mycobactetrium tuberculosis Mfd Oligomer (Dodecamer) / type: protein_or_peptide / ID: 1 / Details: MtbMfd oligomer peak from SEC / Enantiomer: LEVO / EC number: ec: 3.4.6.- |
|---|---|
| Source (natural) | Organism:  Mycobacterium tuberculosis H37Rv (bacteria) Mycobacterium tuberculosis H37Rv (bacteria) |
| Recombinant expression | Organism:  |
| Sequence | String: MGSSHHHHHH SSGLVPRGSH MTAPGPACS DTPIAGLVEL ALSAPTFQQL MQRAGGRPDE LTLIAPASAR LLVASALARQ GPLLVVTATG R EADDLAAE LRGVFGDAVA LLPSWETLPH ERLSPGVDTV GTRLMALRRL AHPDDAQLGP PLGVVVTSVR SL LQPMTPQ ...String: MGSSHHHHHH SSGLVPRGSH MTAPGPACS DTPIAGLVEL ALSAPTFQQL MQRAGGRPDE LTLIAPASAR LLVASALARQ GPLLVVTATG R EADDLAAE LRGVFGDAVA LLPSWETLPH ERLSPGVDTV GTRLMALRRL AHPDDAQLGP PLGVVVTSVR SL LQPMTPQ LGMMEPLTLT VGDESPFDGV VARLVELAYT RVDMVGRRGE FAVRGGILDI FAPTAEHPVR VEF WGDEIT EMRMFSVADQ RSIPEIDIHT LVAFACRELL LSEDVRARAA QLAARHPAAE STVTGSASDM LAKL AEGIA VDGMEAVLPV LWSDGHALLT DQLPDGTPVL VCDPEKVRTR AADLIRTGRE FLEASWSVAA LGTAE NQAP VDVEQLGGSG FVELDQVRAA AARTGHPWWT LSQLSDESAI ELDVRAAPSA RGHQRDIDEI FAMLRA HIA TGGYAALVAP GTGTAHRVVE RLSESDTPAG MLDPGQAPKP GVVGVLQGPL RDGVIIPGAN LVVITET DL TGSRVSAAEG KRLAAKRRNI VDPLALTAGD LVVHDQHGIG RFVEMVERTV GGARREYLVL EYASAKRG G GAKNTDKLYV PMDSLDQLSR YVGGQAPALS RLGGSDWANT KTKARRAVRE IAGELVSLYA KRQASPGHA FSPDTPWQAE LEDAFGFTET VDQLTAIEEV KADMEKPIPM DRVICGDVGY GKTEIAVRAA FKAVQDGKQV AVLVPTTLL ADQHLQTFGE RMSGFPVTIK GLSRFTDAAE SRAVIDGLAD GSVDIVIGTH RLLQTGVRWK D LGLVVVDE EQRFGVEHKE HIKSLRTHVD VLTMSATPIP RTLEMSLAGI REMSTILTPP EERYPVLTYV GP HDDKQIA AALRRELLRD GQAFYVHNRV SSIDAAAARV RELVPEARVV VAHGQMPEDL LETTVQRFWN REH DILVCT TIVETGLDIS NANTLIVERA DTFGLSQLHQ LRGRVGRSRE RGYAYFLYPP QVPLTETAYD RLAT IAQNN ELGAGMAVAL KDLEIRGAGN VLGIEQSGHV AGVGFDLYVR LVGEALETYR DAYRAAADGQ TVRTA EEPK DVRIDLPVDA HLPPDYIASD RLRLEGYRRL AAASSDREVA AVVDELTDRY GALPEPARRL AAVARL RLL CRGSGITDVT AASAATVRLS PLTLPDSAQV RLKRMYPGAH YRATTATVQV PIPRAGGLGA PRIRDVE LV QMVADLITAL AGKPRQHIGI TNPSPPGEDG RGRNTTIKER QP |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 20mM Tris pH8.0, 500mM NaCl, 10mM beta mercaptoethanol |
| Staining | Type: NEGATIVE / Material: Uranyl Acetate Details: Negatively stained EM specimen were prepared by stained with 2% uranyl acetate for one minute and air dried. |
| Grid | Model: Homemade / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS Details: negative stain specimen was prepared by fishing method without Glow Discharge |
| Details | This sample was monodisperse |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 1200EXII |
|---|---|
| Image recording | Film or detector model: GATAN ORIUS SC600 (2.7k x 2.7k) / Number grids imaged: 1 / Number real images: 100 / Average exposure time: 1.0 sec. / Average electron dose: 10.0 e/Å2 / Details: Images were collected on CCD. |
| Electron beam | Acceleration voltage: 100 kV / Electron source: TUNGSTEN HAIRPIN |
| Electron optics | Calibrated magnification: 52974 / Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Cs: 3.5 mm / Nominal magnification: 40000 |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - Chain ID: A |
|---|---|
| Details | Initial manually moved into the EM map and then Rigid body fitted using 43 symmetry 12 mer as single rigid body into the EM map using Chimera. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Correlation coefficient |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)