[English] 日本語
 Yorodumi
Yorodumi- EMDB-9191: Structure of double-stranded target DNA engaged Csy complex from ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9191 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of double-stranded target DNA engaged Csy complex from Pseudomonas aeruginosa (PA-14) | |||||||||
 Map data Map data | Composite sharpened map of the complex generated from multiple focused maps of different sub-regions of the complex. A B-factor of -35 was used for sharpening the map. | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationmaintenance of CRISPR repeat elements / endonuclease activity / defense response to virus / Hydrolases; Acting on ester bonds / RNA binding Similarity search - Function | |||||||||
| Biological species |  Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria) / Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria) /  Pseudomonas aeruginosa UCBPP-PA14 (bacteria) Pseudomonas aeruginosa UCBPP-PA14 (bacteria) | |||||||||
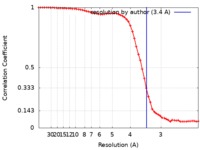
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Chowdhury S / Rollins MF / Carter J / Golden SM / Miettinen HM / Santiago-Frangos A / Faith D / Lawrence MC / Wiedenheft B / Lander GC | |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2019 Journal: Mol Cell / Year: 2019Title: Structure Reveals a Mechanism of CRISPR-RNA-Guided Nuclease Recruitment and Anti-CRISPR Viral Mimicry. Authors: MaryClare F Rollins / Saikat Chowdhury / Joshua Carter / Sarah M Golden / Heini M Miettinen / Andrew Santiago-Frangos / Dominick Faith / C Martin Lawrence / Gabriel C Lander / Blake Wiedenheft /  Abstract: Bacteria and archaea have evolved sophisticated adaptive immune systems that rely on CRISPR RNA (crRNA)-guided detection and nuclease-mediated elimination of invading nucleic acids. Here, we present ...Bacteria and archaea have evolved sophisticated adaptive immune systems that rely on CRISPR RNA (crRNA)-guided detection and nuclease-mediated elimination of invading nucleic acids. Here, we present the cryo-electron microscopy (cryo-EM) structure of the type I-F crRNA-guided surveillance complex (Csy complex) from Pseudomonas aeruginosa bound to a double-stranded DNA target. Comparison of this structure to previously determined structures of this complex reveals a ∼180-degree rotation of the C-terminal helical bundle on the "large" Cas8f subunit. We show that the double-stranded DNA (dsDNA)-induced conformational change in Cas8f exposes a Cas2/3 "nuclease recruitment helix" that is structurally homologous to a virally encoded anti-CRISPR protein (AcrIF3). Structural homology between Cas8f and AcrIF3 suggests that AcrIF3 is a mimic of the Cas8f nuclease recruitment helix. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9191.map.gz emd_9191.map.gz | 11.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9191-v30.xml emd-9191-v30.xml emd-9191.xml emd-9191.xml | 40.5 KB 40.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_9191_fsc.xml emd_9191_fsc.xml | 6.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_9191.png emd_9191.png | 162.4 KB | ||
| Masks |  emd_9191_msk_1.map emd_9191_msk_1.map | 15.6 MB |  Mask map Mask map | |
| Others |  emd_9191_additional_1.map.gz emd_9191_additional_1.map.gz emd_9191_additional_2.map.gz emd_9191_additional_2.map.gz emd_9191_additional_3.map.gz emd_9191_additional_3.map.gz emd_9191_additional_4.map.gz emd_9191_additional_4.map.gz emd_9191_additional_5.map.gz emd_9191_additional_5.map.gz emd_9191_additional_6.map.gz emd_9191_additional_6.map.gz emd_9191_half_map_1.map.gz emd_9191_half_map_1.map.gz emd_9191_half_map_2.map.gz emd_9191_half_map_2.map.gz | 13.2 MB 14.5 MB 14.2 MB 13.5 MB 13.3 MB 13.7 MB 13.6 MB 13.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9191 http://ftp.pdbj.org/pub/emdb/structures/EMD-9191 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9191 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9191 | HTTPS FTP |
-Related structure data
| Related structure data |  6ne0MC  6mpu M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9191.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9191.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite sharpened map of the complex generated from multiple focused maps of different sub-regions of the complex. A B-factor of -35 was used for sharpening the map. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
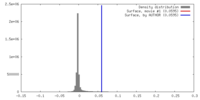
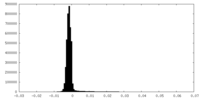
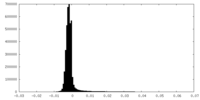
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_9191_msk_1.map emd_9191_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
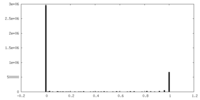
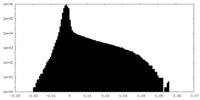
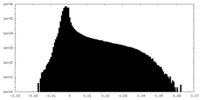
| Density Histograms |
-Additional map: Composite unsharpened map of the complex generated from...
| File | emd_9191_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Composite unsharpened map of the complex generated from multiple focused maps of different sub-regions of the complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
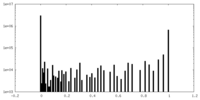
| Density Histograms |
-Additional map: Sharpened non-focused map of the full complex
| File | emd_9191_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened non-focused map of the full complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
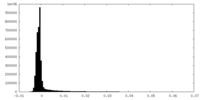
| Density Histograms |
-Additional map: Unsharpened non-focused map of the full complex
| File | emd_9191_additional_3.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unsharpened non-focused map of the full complex | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Focused map of "Region-1" of the Csy-DNA complex,...
| File | emd_9191_additional_4.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused map of "Region-1" of the Csy-DNA complex, comprising the head (Cas6f-crRNA stem loop) - Cas8f C-terminal helix bundle - Cas7.1f and Cas7.2f subunits | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Focused map of "Region-2" of the Csy-DNA complex,...
| File | emd_9191_additional_5.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused map of "Region-2" of the Csy-DNA complex, comprising all six Cas7f subunits - crRNA - complimentary target DNA strand | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: Focused map of "Region-3" of the Csy-DNA complex,...
| File | emd_9191_additional_6.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Focused map of "Region-3" of the Csy-DNA complex, comprising the tail (Cas8f N-terminal half - Cas5f - double-stranded target DNA) - Cas7.6 subunit | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered composite half map #1
| File | emd_9191_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered composite half map #1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Unfiltered composite half map #2
| File | emd_9191_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered composite half map #2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Double-stranded target DNA engaged Csy Complex from Pseudomonas a...
| Entire | Name: Double-stranded target DNA engaged Csy Complex from Pseudomonas aeruginosa (PA-14) |
|---|---|
| Components |
|
-Supramolecule #1: Double-stranded target DNA engaged Csy Complex from Pseudomonas a...
| Supramolecule | Name: Double-stranded target DNA engaged Csy Complex from Pseudomonas aeruginosa (PA-14) type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#7 |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria) Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria) |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 360 KDa |
-Macromolecule #1: CRISPR-associated protein Csy2
| Macromolecule | Name: CRISPR-associated protein Csy2 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria) Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria)Strain: UCBPP-PA14 |
| Molecular weight | Theoretical: 36.244074 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSVTDPEALL LLPRLSIQNA NAISSPLTWG FPSPGAFTGF VHALQRRVGI SLDIELDGVG IVCHRFEAQI SQPAGKRTKV FNLTRNPLN RDGSTAAIVE EGRAHLEVSL LLGVHGDGLD DHPAQEIARQ VQEQAGAMRL AGGSILPWCN ERFPAPNAEL L MLGGSDEQ ...String: MSVTDPEALL LLPRLSIQNA NAISSPLTWG FPSPGAFTGF VHALQRRVGI SLDIELDGVG IVCHRFEAQI SQPAGKRTKV FNLTRNPLN RDGSTAAIVE EGRAHLEVSL LLGVHGDGLD DHPAQEIARQ VQEQAGAMRL AGGSILPWCN ERFPAPNAEL L MLGGSDEQ RRKNQRRLTR RLLPGFALVS REALLQQHLE TLRTTLPEAT TLDALLDLCR INFEPPATSS EEEASPPDAA WQ VRDKPGW LVPIPAGYNA LSPLYLPGEV RNARDRETPL RFVENLFGLG EWLSPHRVAA LSDLLWYHHA EPDKGLYRWS TPR FVEHAI A |
-Macromolecule #2: CRISPR-associated protein Csy3
| Macromolecule | Name: CRISPR-associated protein Csy3 / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria) Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria)Strain: UCBPP-PA14 |
| Molecular weight | Theoretical: 37.579273 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSKPILSTAS VLAFERKLDP SDALMSAGAW AQRDASQEWP AVTVREKSVR GTISNRLKTK DRDPAKLDAS IQSPNLQTVD VANLPSDAD TLKVRFTLRV LGGAGTPSAC NDAAYRDKLL QTVATYVNDQ GFAELARRYA HNLANARFLW RNRVGAEAVE V RINHIRQG ...String: MSKPILSTAS VLAFERKLDP SDALMSAGAW AQRDASQEWP AVTVREKSVR GTISNRLKTK DRDPAKLDAS IQSPNLQTVD VANLPSDAD TLKVRFTLRV LGGAGTPSAC NDAAYRDKLL QTVATYVNDQ GFAELARRYA HNLANARFLW RNRVGAEAVE V RINHIRQG EVARAWRFDA LAIGLRDFKA DAELDALAEL IASGLSGSGH VLLEVVAFAR IGDGQEVFPS QELILDKGDK KG QKSKTLY SVRDAAAIHS QKIGNALRTI DTWYPDEDGL GPIAVEPYGS VTSQGKAYRQ PKQKLDFYTL LDNWVLRDEA PAV EQQHYV IANLIRGGVF GEAEEK |
-Macromolecule #3: CRISPR-associated endonuclease Cas6/Csy4
| Macromolecule | Name: CRISPR-associated endonuclease Cas6/Csy4 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: Hydrolases; Acting on ester bonds |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria) Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria)Strain: UCBPP-PA14 |
| Molecular weight | Theoretical: 21.675781 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: FTMDHYLDIR LRPDPEFPPA QLMSVLFGKL HQALVAQGGD RIGVSFPDLD ESRSRLGERL RIHASADDLR ALLARPWLEG LRDHLQFGE PAVVPHPTPY RQVSRVQAKS NPERLRRRLM RRHDLSEEEA RKRIPDTVAR ALDLPFVTLR SQSTGQHFRL F IRHGPLQV TAEEGGFTCY GLSKGGFVPW F |
-Macromolecule #7: CRISPR-associated protein Csy1
| Macromolecule | Name: CRISPR-associated protein Csy1 / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria) Pseudomonas aeruginosa (strain UCBPP-PA14) (bacteria)Strain: UCBPP-PA14 |
| Molecular weight | Theoretical: 49.194168 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTSPLPTPTW QELRQFIESF IQERLQGKLD KLQPDEDDKR QTLLATHRRE AWLADAARRV GQLQLVTHTL KPIHPDARGS NLHSLPQAP GQPGLAGSHE LGDRLVSDVV GNAAALDVFK FLSLQYQGKN LLNWLTEDSA EALQALSDNA EQAREWRQAF I GITTVKGA ...String: MTSPLPTPTW QELRQFIESF IQERLQGKLD KLQPDEDDKR QTLLATHRRE AWLADAARRV GQLQLVTHTL KPIHPDARGS NLHSLPQAP GQPGLAGSHE LGDRLVSDVV GNAAALDVFK FLSLQYQGKN LLNWLTEDSA EALQALSDNA EQAREWRQAF I GITTVKGA PASHSLAKQL YFPLPGSGYH LLAPLFPTSL VHHVHALLRE ARFGDAAKAA REARSRQESW PHGFSEYPNL AI QKFGGTK PQNISQLNNE RRGENWLLPS LPPNWQRQNV NAPMRHSSVF EHDFGRTPEV SRLTRTLQRF LAKTVHNNLA IRQ RRAQLV AQICDEALQY AARLRELEPG WSATPGCQLH DAEQLWLDPL RAQTDETFLQ RRLRGDWPAE VGNRFANWLN RAVS SDSQI LGSPEAAQWS QELSKELTMF KEILEDERD |
-Macromolecule #4: CRISPR RNA (60-MER)
| Macromolecule | Name: CRISPR RNA (60-MER) / type: rna / ID: 4 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa UCBPP-PA14 (bacteria) Pseudomonas aeruginosa UCBPP-PA14 (bacteria) |
| Molecular weight | Theoretical: 19.265404 KDa |
| Sequence | String: CUAAGAAAUU CACGGCGGGC UUGAUGUCCG CGUCUACCUG GUUCACUGCC GUGUAGGCAG |
-Macromolecule #5: CRISPR target DNA (44-MER)
| Macromolecule | Name: CRISPR target DNA (44-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa UCBPP-PA14 (bacteria) Pseudomonas aeruginosa UCBPP-PA14 (bacteria) |
| Molecular weight | Theoretical: 13.584703 KDa |
| Sequence | String: (DC)(DA)(DG)(DG)(DT)(DA)(DG)(DA)(DC)(DG) (DC)(DG)(DG)(DA)(DC)(DA)(DT)(DC)(DA)(DA) (DG)(DC)(DC)(DC)(DG)(DC)(DC)(DG)(DT) (DG)(DA)(DA)(DG)(DG)(DT)(DG)(DC)(DA)(DG) (DC) (DT)(DT)(DC)(DT) |
-Macromolecule #6: Non-complementary R-loop DNA strand
| Macromolecule | Name: Non-complementary R-loop DNA strand / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism:  Pseudomonas aeruginosa UCBPP-PA14 (bacteria) Pseudomonas aeruginosa UCBPP-PA14 (bacteria) |
| Molecular weight | Theoretical: 10.476714 KDa |
| Sequence | String: (DA)(DG)(DA)(DA)(DG)(DC)(DT)(DG)(DC)(DA) (DC)(DC)(DT)(DT)(DC)(DA)(DC)(DG)(DG)(DC) (DG)(DG)(DG)(DC)(DT)(DT)(DG)(DA)(DT) (DG)(DT)(DC)(DC)(DG) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Atmosphere: OTHER | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER Details: Freezing was carried out in a cold room at 4 degrees C and relative humidity of 98%. 5 uL sample was applied to plasma cleaned grid and manually blotted with Whatman 1 filter paper for 5-7 ...Details: Freezing was carried out in a cold room at 4 degrees C and relative humidity of 98%. 5 uL sample was applied to plasma cleaned grid and manually blotted with Whatman 1 filter paper for 5-7 sec before plunge freezing.. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Temperature | Min: 77.0 K / Max: 79.0 K |
| Details | Objective astigmatism was corrected at 36000x magnification using Thon rings visualized with a K2 camera. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Dimensions - Width: 7676 pixel / Digitization - Dimensions - Height: 7420 pixel / Digitization - Sampling interval: 2.5 µm / Digitization - Frames/image: 1-56 / Number grids imaged: 4 / Number real images: 3208 / Average exposure time: 14.0 sec. / Average electron dose: 58.0 e/Å2 Details: Data were acquired using Leginon and collected on K2 summit operating in super-resolution mode. |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: |
|---|---|
| Details | The atomic models for Cas5f, Cas8f, Cas6f, and Cas7f from the Csy Acr complex (PDB ID 5UZ9) were used as initial template models for model building. These were individually rigid body-fitted into the reconstructed maps using the fit map function in UCSF Chimera, and residue registers and backbone geometries were fixed in Coot. Models for the crRNA and DNA strands were also manually built into the map using Coot. |
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-6ne0: |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)