+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-9013 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | EsCas13d-crRNA binary complex | |||||||||
 Map data Map data | EsCas13d-crRNA binary complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CRISPR-Cas / RNase / Complex / RNA BINDING PROTEIN-RNA complex | |||||||||
| Function / homology | Uncharacterized protein Function and homology information Function and homology information | |||||||||
| Biological species |  unidentified bacterium (bacteria) / unidentified bacterium (bacteria) /  bacterium (bacteria) / bacterium (bacteria) /  [Eubacterium] siraeum DSM 15702 (bacteria) [Eubacterium] siraeum DSM 15702 (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.4 Å | |||||||||
 Authors Authors | Zhang C / Lyumkis D | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2018 Journal: Cell / Year: 2018Title: Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d. Authors: Cheng Zhang / Silvana Konermann / Nicholas J Brideau / Peter Lotfy / Xuebing Wu / Scott J Novick / Timothy Strutzenberg / Patrick R Griffin / Patrick D Hsu / Dmitry Lyumkis /  Abstract: CRISPR-Cas endonucleases directed against foreign nucleic acids mediate prokaryotic adaptive immunity and have been tailored for broad genetic engineering applications. Type VI-D CRISPR systems ...CRISPR-Cas endonucleases directed against foreign nucleic acids mediate prokaryotic adaptive immunity and have been tailored for broad genetic engineering applications. Type VI-D CRISPR systems contain the smallest known family of single effector Cas enzymes, and their signature Cas13d ribonuclease employs guide RNAs to cleave matching target RNAs. To understand the molecular basis for Cas13d function and explain its compact molecular architecture, we resolved cryoelectron microscopy structures of Cas13d-guide RNA binary complex and Cas13d-guide-target RNA ternary complex to 3.4 and 3.3 Å resolution, respectively. Furthermore, a 6.5 Å reconstruction of apo Cas13d combined with hydrogen-deuterium exchange revealed conformational dynamics that have implications for RNA scanning. These structures, together with biochemical and cellular characterization, provide insights into its RNA-guided, RNA-targeting mechanism and delineate a blueprint for the rational design of improved transcriptome engineering technologies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_9013.map.gz emd_9013.map.gz | 56.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-9013-v30.xml emd-9013-v30.xml emd-9013.xml emd-9013.xml | 22.2 KB 22.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_9013.png emd_9013.png | 173.9 KB | ||
| Filedesc metadata |  emd-9013.cif.gz emd-9013.cif.gz | 6.9 KB | ||
| Others |  emd_9013_additional.map.gz emd_9013_additional.map.gz emd_9013_half_map_1.map.gz emd_9013_half_map_1.map.gz emd_9013_half_map_2.map.gz emd_9013_half_map_2.map.gz | 5.8 MB 11.8 MB 11.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-9013 http://ftp.pdbj.org/pub/emdb/structures/EMD-9013 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9013 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-9013 | HTTPS FTP |
-Validation report
| Summary document |  emd_9013_validation.pdf.gz emd_9013_validation.pdf.gz | 799.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_9013_full_validation.pdf.gz emd_9013_full_validation.pdf.gz | 798.7 KB | Display | |
| Data in XML |  emd_9013_validation.xml.gz emd_9013_validation.xml.gz | 12.4 KB | Display | |
| Data in CIF |  emd_9013_validation.cif.gz emd_9013_validation.cif.gz | 14.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9013 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9013 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9013 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-9013 | HTTPS FTP |
-Related structure data
| Related structure data |  6e9eMC  9014C  9015C  6e9fC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_9013.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_9013.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EsCas13d-crRNA binary complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.79 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
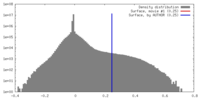
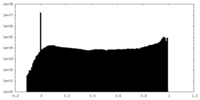
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Additional map: 3D FSC map
| File | emd_9013_additional.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | 3D FSC map | ||||||||||||
| Projections & Slices |
| ||||||||||||
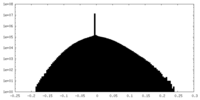
| Density Histograms |
-Half map: EsCas13d-crRNA binary complex, half map #1
| File | emd_9013_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EsCas13d-crRNA binary complex, half map #1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
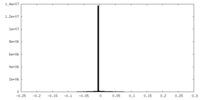
| Density Histograms |
-Half map: EsCas13d-crRNA binary complex, half map #2
| File | emd_9013_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | EsCas13d-crRNA binary complex, half map #2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
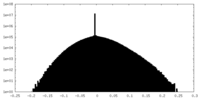
| Density Histograms |
- Sample components
Sample components
-Entire : Binary complex of EsCas13d with crRNA and Mg2+
| Entire | Name: Binary complex of EsCas13d with crRNA and Mg2+ |
|---|---|
| Components |
|
-Supramolecule #1: Binary complex of EsCas13d with crRNA and Mg2+
| Supramolecule | Name: Binary complex of EsCas13d with crRNA and Mg2+ / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  unidentified bacterium (bacteria) unidentified bacterium (bacteria) |
| Molecular weight | Theoretical: 125 KDa |
-Macromolecule #1: crRNA (52-MER)
| Macromolecule | Name: crRNA (52-MER) / type: rna / ID: 1 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  bacterium (bacteria) bacterium (bacteria) |
| Molecular weight | Theoretical: 16.385846 KDa |
| Sequence | String: CACCCGUGCA AAAAUGCAGG GGUCUAAAAC GACCUGAAUA UUUCAGAUCA A |
-Macromolecule #2: EsCas13d
| Macromolecule | Name: EsCas13d / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  [Eubacterium] siraeum DSM 15702 (bacteria) [Eubacterium] siraeum DSM 15702 (bacteria) |
| Molecular weight | Theoretical: 110.828938 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGKKIHARDL REQRKTDRTE KFADQNKKRE AERAVPKKDA AVSVKSVSSV SSKKDNVTKS MAKAAGVKSV FAVGNTVYMT SFGRGNDAV LEQKIVDTSH EPLNIDDPAY QLNVVTMNGY SVTGHRGETV SAVTDNPLRR FNGRKKDEPE QSVPTDMLCL K PTLEKKFF ...String: MGKKIHARDL REQRKTDRTE KFADQNKKRE AERAVPKKDA AVSVKSVSSV SSKKDNVTKS MAKAAGVKSV FAVGNTVYMT SFGRGNDAV LEQKIVDTSH EPLNIDDPAY QLNVVTMNGY SVTGHRGETV SAVTDNPLRR FNGRKKDEPE QSVPTDMLCL K PTLEKKFF GKEFDDNIHI QLIYNILDIE KILAVYSTNA IYALNNMSAD ENIENSDFFM KRTTDETFDD FEKKKESTNS RE KADFDAF EKFIGNYRLA YFADAFYVNK KNPKGKAKNV LREDKELYSV LTLIGKLRHW CVHSEEGRAE FWLYKLDELK DDF KNVLDV VYNRPVEEIN NRFIENNKVN IQILGSVYKN TDIAELVRSY YEFLITKKYK NMGFSIKKLR ESMLEGKGYA DKEY DSVRN KLYQMTDFIL YTGYINEDSD RADDLVNTLR SSLKEDDKTT VYCKEADYLW KKYRESIREV ADALDGDNIK KLSKS NIEI QEDKLRKCFI SYADSVSEFT KLIYLLTRFL SGKEINDLVT TLINKFDNIR SFLEIMDELG LDRTFTAEYS FFEGST KYL AELVELNSFV KSCSFDINAK RTMYRDALDI LGIESDKTEE DIEKMIDNIL QIDANGDKKL KKNNGLRNFI ASNVIDS NR FKYLVRYGNP KKIRETAKCK PAVRFVLNEI PDAQIERYYE ACCPKNTALC SANKRREKLA DMIAEIKFEN FSDAGNYQ K ANVTSRTSEA EIKRKNQAII RLYLTVMYIM LKNLVNVNAR YVIAFHCVER DTKLYAESGL EVGNIEKNKT NLTMAVMGV KLENGIIKTE FDKSFAENAA NRYLRNARWY KLILDNLKKS ERAVVNEFRN TVCHLNAIRN ININIKEIKE VENYFALYHY LIQKHLENR FADKKVERDT GDFISKLEEH KTYCKDFVKA YCTPFGYNLV RYKNLTIDGL FDKNYPGKDD SDEQK UniProtKB: Uncharacterized protein |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 1 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1 mg/mL | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||||||||||||||
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 7 sec. / Pretreatment - Atmosphere: OTHER | |||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-120 / Number real images: 1435 / Average electron dose: 56.8 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 57000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Algorithm: FOURIER SPACE / Resolution.type: BY AUTHOR / Resolution: 3.4 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cisTEM / Number images used: 43786 |
| Initial angle assignment | Type: PROJECTION MATCHING / Software - Name: RELION |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name: cisTEM |
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: AB INITIO MODEL / Target criteria: Correlation coefficient |
|---|---|
| Output model |  PDB-6e9e: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)