[English] 日本語
 Yorodumi
Yorodumi- PDB-8f0q: Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the acyl... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 8f0q | ||||||
|---|---|---|---|---|---|---|---|
| Title | Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the acylsulfonamide inhibitor GDC-0310 | ||||||
 Components Components | Sodium channel protein PaFPC1,Sodium channel protein type 9 subunit alpha chimera | ||||||
 Keywords Keywords | MEMBRANE PROTEIN/INHIBITOR / Ion channel / small molecule / inhibitor / MEMBRANE PROTEIN-INHIBITOR complex | ||||||
| Function / homology |  Function and homology information Function and homology informationaction potential propagation / detection of mechanical stimulus involved in sensory perception / membrane depolarization during action potential / cardiac muscle cell action potential involved in contraction / node of Ranvier / voltage-gated sodium channel complex / voltage-gated monoatomic cation channel activity / Interaction between L1 and Ankyrins / voltage-gated sodium channel activity / detection of temperature stimulus involved in sensory perception of pain ...action potential propagation / detection of mechanical stimulus involved in sensory perception / membrane depolarization during action potential / cardiac muscle cell action potential involved in contraction / node of Ranvier / voltage-gated sodium channel complex / voltage-gated monoatomic cation channel activity / Interaction between L1 and Ankyrins / voltage-gated sodium channel activity / detection of temperature stimulus involved in sensory perception of pain / Phase 0 - rapid depolarisation / behavioral response to pain / neuronal action potential / sensory perception of pain / axon terminus / sodium ion transmembrane transport / post-embryonic development / circadian rhythm / response to toxic substance / Sensory perception of sweet, bitter, and umami (glutamate) taste / inflammatory response / axon / plasma membrane Similarity search - Function | ||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) Periplaneta americana (American cockroach) Periplaneta americana (American cockroach) | ||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 2.5 Å | ||||||
 Authors Authors | Kschonsak, M. / Jao, C.C. / Arthur, C.P. / Rohou, A.L. / Bergeron, P. / Ortwine, D. / McKerall, S.J. / Hackos, D.H. / Deng, L. / Chen, J. ...Kschonsak, M. / Jao, C.C. / Arthur, C.P. / Rohou, A.L. / Bergeron, P. / Ortwine, D. / McKerall, S.J. / Hackos, D.H. / Deng, L. / Chen, J. / Sutherlin, D. / Dragovich, P.S. / Volgraf, M. / Wright, M.R. / Payandeh, J. / Ciferri, C. / Tellis, J.C. | ||||||
| Funding support | 1items
| ||||||
 Citation Citation |  Journal: Elife / Year: 2023 Journal: Elife / Year: 2023Title: Cryo-EM reveals an unprecedented binding site for Na1.7 inhibitors enabling rational design of potent hybrid inhibitors. Authors: Marc Kschonsak / Christine C Jao / Christopher P Arthur / Alexis L Rohou / Philippe Bergeron / Daniel F Ortwine / Steven J McKerrall / David H Hackos / Lunbin Deng / Jun Chen / Tianbo Li / ...Authors: Marc Kschonsak / Christine C Jao / Christopher P Arthur / Alexis L Rohou / Philippe Bergeron / Daniel F Ortwine / Steven J McKerrall / David H Hackos / Lunbin Deng / Jun Chen / Tianbo Li / Peter S Dragovich / Matthew Volgraf / Matthew R Wright / Jian Payandeh / Claudio Ciferri / John C Tellis /  Abstract: The voltage-gated sodium (Na) channel Na1.7 has been identified as a potential novel analgesic target due to its involvement in human pain syndromes. However, clinically available Na channel-blocking ...The voltage-gated sodium (Na) channel Na1.7 has been identified as a potential novel analgesic target due to its involvement in human pain syndromes. However, clinically available Na channel-blocking drugs are not selective among the nine Na channel subtypes, Na1.1-Na1.9. Moreover, the two currently known classes of Na1.7 subtype-selective inhibitors (aryl- and acylsulfonamides) have undesirable characteristics that may limit their development. To this point understanding of the structure-activity relationships of the acylsulfonamide class of Na1.7 inhibitors, exemplified by the clinical development candidate , has been based solely on a single co-crystal structure of an arylsulfonamide inhibitor bound to voltage-sensing domain 4 (VSD4). To advance inhibitor design targeting the Na1.7 channel, we pursued high-resolution ligand-bound Na1.7-VSD4 structures using cryogenic electron microscopy (cryo-EM). Here, we report that engages the Na1.7-VSD4 through an unexpected binding mode orthogonal to the arylsulfonamide inhibitor class binding pose, which identifies a previously unknown ligand binding site in Na channels. This finding enabled the design of a novel hybrid inhibitor series that bridges the aryl- and acylsulfonamide binding pockets and allows for the generation of molecules with substantially differentiated structures and properties. Overall, our study highlights the power of cryo-EM methods to pursue challenging drug targets using iterative and high-resolution structure-guided inhibitor design. This work also underscores an important role of the membrane bilayer in the optimization of selective Na channel modulators targeting VSD4. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  8f0q.cif.gz 8f0q.cif.gz | 250.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb8f0q.ent.gz pdb8f0q.ent.gz | 194.9 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  8f0q.json.gz 8f0q.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/f0/8f0q https://data.pdbj.org/pub/pdb/validation_reports/f0/8f0q ftp://data.pdbj.org/pub/pdb/validation_reports/f0/8f0q ftp://data.pdbj.org/pub/pdb/validation_reports/f0/8f0q | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  28777MC  8f0pC  8f0rC  8f0sC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 184481.906 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Details: Chimeric construct of human Nav1.7 VSD4 and the NavPaS channel from American cockroach Periplaneta americana Source: (gene. exp.)  Homo sapiens (human), (gene. exp.) Homo sapiens (human), (gene. exp.)  Periplaneta americana (American cockroach) Periplaneta americana (American cockroach)Gene: SCN9A, NENA / Cell line (production host): 293 suspension cells / Production host:  Homo sapiens (human) / References: UniProt: D0E0C2, UniProt: Q15858 Homo sapiens (human) / References: UniProt: D0E0C2, UniProt: Q15858 |
|---|
-Sugars , 3 types, 4 molecules 


| #2: Polysaccharide | beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3) ...beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose Type: oligosaccharide / Mass: 748.682 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source |
|---|---|
| #3: Sugar | ChemComp-BMA / |
| #4: Sugar |
-Non-polymers , 4 types, 85 molecules 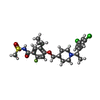






| #5: Chemical | ChemComp-X7R / | ||||
|---|---|---|---|---|---|
| #6: Chemical | ChemComp-PEE / #7: Chemical | ChemComp-Y01 / | #8: Water | ChemComp-HOH / | |
-Details
| Has ligand of interest | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Structure of VSD4-NaV1.7-NaVPas channel chimera bound to the acylsulfonamide inhibitor GDC-0310 Type: COMPLEX Details: Chimeric construct of human Nav1.7 VSD4 and the NavPaS channel from American cockroach Periplaneta americana Entity ID: #1 / Source: MULTIPLE SOURCES | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular weight | Experimental value: NO | |||||||||||||||
| Source (natural) |
| |||||||||||||||
| Source (recombinant) | Organism:  Homo sapiens (human) / Cell: 293 suspension cells Homo sapiens (human) / Cell: 293 suspension cells | |||||||||||||||
| Buffer solution | pH: 7.5 | |||||||||||||||
| Buffer component |
| |||||||||||||||
| Specimen | Conc.: 2 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES Details: The sample was reconstituted into lipid nanodiscs (MSP1E3D1 in 3POPC:1POPE:1POPG) and was monodisperse. The sample was crosslinked with 0.05% glutaraldehyde for 10 minutes at RT, then ...Details: The sample was reconstituted into lipid nanodiscs (MSP1E3D1 in 3POPC:1POPE:1POPG) and was monodisperse. The sample was crosslinked with 0.05% glutaraldehyde for 10 minutes at RT, then quenched with 1M Tris pH7.0. | |||||||||||||||
| Specimen support | Details: Grids were incubated with a thiol reactive, self-assembling reaction mixture of 4mM monothiolalkane(C11)PEG6-OH (11-mercaptoundecyl) hexaethyleneglycol (SPT-0011P6, SensoPath Technologies, ...Details: Grids were incubated with a thiol reactive, self-assembling reaction mixture of 4mM monothiolalkane(C11)PEG6-OH (11-mercaptoundecyl) hexaethyleneglycol (SPT-0011P6, SensoPath Technologies, Inc., Bozeman, MT). Grids were incubated with this self-assembled monolayer (SAM) solution for 24 hours and afterwards rinsed with EtOH. Grid material: GOLD / Grid mesh size: 300 divisions/in. / Grid type: UltrAuFoil R0./1 | |||||||||||||||
| Vitrification | Instrument: FEI VITROBOT MARK IV / Cryogen name: ETHANE / Humidity: 100 % / Chamber temperature: 277.15 K |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD / Nominal defocus max: 1500 nm / Nominal defocus min: 500 nm |
| Image recording | Average exposure time: 3 sec. / Electron dose: 60 e/Å2 / Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) |
- Processing
Processing
| EM software |
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTF correction | Details: microgaphs with CTFfit of 6.0 A or better were selected Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||
| Particle selection | Num. of particles selected: 2253983 Details: template-matching particle picking with a 30A low-pass filtered template | ||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||||||||||||||||||||||||||||||
| 3D reconstruction | Resolution: 2.5 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 795792 Details: A score threshold was applied, so that only the best-scoring particle images would be included in the 3D reconstruction at each cycle. Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: FLEXIBLE FIT / Space: REAL |
 Movie
Movie Controller
Controller




 PDBj
PDBj










