+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8911 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of a mitochondrial calcium uniporter | |||||||||
 Map data Map data | Single-particle cryo-EM reconstruction of the N. crassa mitochondrial calcium uniporter | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | mitochondrial calcium uniporter / calcium-selective ion channel / calcium uptake / uniporter / TRANSPORT PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationuniporter activity / uniplex complex / mitochondrial calcium ion homeostasis / calcium import into the mitochondrion / calcium channel activity / protein homotetramerization / mitochondrial inner membrane / metal ion binding / identical protein binding Similarity search - Function | |||||||||
| Biological species |  Neurospora crassa (fungus) Neurospora crassa (fungus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Yoo J / Wu M | |||||||||
 Citation Citation |  Journal: Science / Year: 2018 Journal: Science / Year: 2018Title: Cryo-EM structure of a mitochondrial calcium uniporter. Authors: Jiho Yoo / Mengyu Wu / Ying Yin / Mark A Herzik / Gabriel C Lander / Seok-Yong Lee /  Abstract: Calcium transport plays an important role in regulating mitochondrial physiology and pathophysiology. The mitochondrial calcium uniporter (MCU) is a calcium-selective ion channel that is the primary ...Calcium transport plays an important role in regulating mitochondrial physiology and pathophysiology. The mitochondrial calcium uniporter (MCU) is a calcium-selective ion channel that is the primary mediator for calcium uptake into the mitochondrial matrix. Here, we present the cryo-electron microscopy structure of the full-length MCU from to an overall resolution of ~3.7 angstroms. Our structure reveals a tetrameric architecture, with the soluble and transmembrane domains adopting different symmetric arrangements within the channel. The conserved W-D-Φ-Φ-E-P-V-T-Y sequence motif of MCU pore forms a selectivity filter comprising two acidic rings separated by one helical turn along the central axis of the channel pore. The structure combined with mutagenesis gives insight into the basis of calcium recognition. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8911.map.gz emd_8911.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8911-v30.xml emd-8911-v30.xml emd-8911.xml emd-8911.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_8911_fsc.xml emd_8911_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_8911.png emd_8911.png | 155.8 KB | ||
| Filedesc metadata |  emd-8911.cif.gz emd-8911.cif.gz | 5.7 KB | ||
| Others |  emd_8911_half_map_1.map.gz emd_8911_half_map_1.map.gz emd_8911_half_map_2.map.gz emd_8911_half_map_2.map.gz | 48.3 MB 48.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8911 http://ftp.pdbj.org/pub/emdb/structures/EMD-8911 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8911 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8911 | HTTPS FTP |
-Related structure data
| Related structure data |  6dt0MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_8911.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8911.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Single-particle cryo-EM reconstruction of the N. crassa mitochondrial calcium uniporter | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.15 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: N. crassa mitochondrial calcium uniporter, half map 1
| File | emd_8911_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | N. crassa mitochondrial calcium uniporter, half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
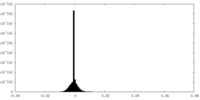
| Density Histograms |
-Half map: N. crassa mitochondrial calcium uniporter, half map 2
| File | emd_8911_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | N. crassa mitochondrial calcium uniporter, half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
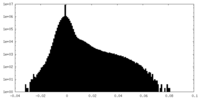
| Density Histograms |
- Sample components
Sample components
-Entire : Mitochondrial Calcium Uniporter
| Entire | Name: Mitochondrial Calcium Uniporter |
|---|---|
| Components |
|
-Supramolecule #1: Mitochondrial Calcium Uniporter
| Supramolecule | Name: Mitochondrial Calcium Uniporter / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
-Macromolecule #1: Mitochondrial calcium uniporter
| Macromolecule | Name: Mitochondrial calcium uniporter / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Neurospora crassa (fungus) Neurospora crassa (fungus) |
| Molecular weight | Theoretical: 52.948117 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASTSVSPKT RETEAEAKAK KLDQKRLDEH EEEVRAREQQ VRRPWHREGA DKPPVEGNAD PIAKGKLLTT PTRLLKLILP LPLRVEKDQ KNNGRNNEYG RSISLNSDIQ PLALLIHPQQ PLSYVERLIQ AELPPVVENG QEKIPNVYFR AEDSEQGDQK P TSRAEARS ...String: MASTSVSPKT RETEAEAKAK KLDQKRLDEH EEEVRAREQQ VRRPWHREGA DKPPVEGNAD PIAKGKLLTT PTRLLKLILP LPLRVEKDQ KNNGRNNEYG RSISLNSDIQ PLALLIHPQQ PLSYVERLIQ AELPPVVENG QEKIPNVYFR AEDSEQGDQK P TSRAEARS KDDGGEPSEY NTNLSHVASA SGLGHRGPKR SSQDKRWVRW SSSTEMGDFI RDAARGREFA IEIEGYNIEM RV SVPSFGD RTYYMRQRLR KMSSEIDGLA KIKHECDLLA HRSAHRLAKG GFGLLAGWWG VVYYVTFHTE FGWDLVEPVT YLA GLTTIM GGYLWFLYIN KDLSYKAAMN VTVSRRQHAL YEMKGFDIER WEQLVQDANA LRREIRVIAV EYDVDWDETR DVGE DVKDV LDEERSRRDD EHRSIEKEKD EKFTEDEKRK RKKDKESKET SGDSTNSHHH HHH UniProtKB: Calcium uniporter protein, mitochondrial |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 1 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.2 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 7 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: HOMEMADE PLUNGER / Details: 4 second blot time. |
| Details | Final reconstruction comprises ~80% of MCU in nanodisc (crosslinked using BS3) and ~20% MCU in amphipol. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3710 pixel / Digitization - Dimensions - Height: 3838 pixel / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 36000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Overall B value: 100 |
|---|---|
| Output model |  PDB-6dt0: |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)