+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-8300 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Casp-8 tDED filament (CASP target) | |||||||||
 Map data Map data | Casp-8 tDED filament | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationcaspase-8 / syncytiotrophoblast cell differentiation involved in labyrinthine layer development / death effector domain binding / FasL/ CD95L signaling / TRAIL signaling / CD95 death-inducing signaling complex / Apoptotic execution phase / ripoptosome / Defective RIPK1-mediated regulated necrosis / Activation, myristolyation of BID and translocation to mitochondria ...caspase-8 / syncytiotrophoblast cell differentiation involved in labyrinthine layer development / death effector domain binding / FasL/ CD95L signaling / TRAIL signaling / CD95 death-inducing signaling complex / Apoptotic execution phase / ripoptosome / Defective RIPK1-mediated regulated necrosis / Activation, myristolyation of BID and translocation to mitochondria / Microbial modulation of RIPK1-mediated regulated necrosis / TRAIL-activated apoptotic signaling pathway / TRIF-mediated programmed cell death / Regulation by c-FLIP / CASP8 activity is inhibited / Dimerization of procaspase-8 / TLR3-mediated TICAM1-dependent programmed cell death / Caspase activation via Death Receptors in the presence of ligand / self proteolysis / positive regulation of macrophage differentiation / response to cobalt ion / NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 / CLEC7A/inflammasome pathway / death-inducing signaling complex / negative regulation of necroptotic process / regulation of tumor necrosis factor-mediated signaling pathway / tumor necrosis factor receptor binding / death receptor binding / natural killer cell activation / TNFR1-induced proapoptotic signaling / RIPK1-mediated regulated necrosis / response to anesthetic / execution phase of apoptosis / regulation of innate immune response / Apoptotic cleavage of cellular proteins / pyroptotic inflammatory response / response to tumor necrosis factor / B cell activation / positive regulation of proteolysis / macrophage differentiation / extrinsic apoptotic signaling pathway via death domain receptors / positive regulation of execution phase of apoptosis / Caspase-mediated cleavage of cytoskeletal proteins / extrinsic apoptotic signaling pathway / negative regulation of canonical NF-kappaB signal transduction / cysteine-type peptidase activity / regulation of cytokine production / proteolysis involved in protein catabolic process / T cell activation / protein maturation / positive regulation of interleukin-1 beta production / Regulation of NF-kappa B signaling / apoptotic signaling pathway / Regulation of TNFR1 signaling / cellular response to mechanical stimulus / NOD1/2 Signaling Pathway / protein processing / Regulation of necroptotic cell death / response to estradiol / peptidase activity / positive regulation of neuron apoptotic process / lamellipodium / heart development / cell body / scaffold protein binding / response to ethanol / angiogenesis / response to lipopolysaccharide / mitochondrial outer membrane / cytoskeleton / positive regulation of canonical NF-kappaB signal transduction / positive regulation of cell migration / positive regulation of apoptotic process / cysteine-type endopeptidase activity / apoptotic process / ubiquitin protein ligase binding / protein-containing complex binding / protein-containing complex / mitochondrion / proteolysis / identical protein binding / nucleus / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 4.6 Å | |||||||||
 Authors Authors | Fu TM / Li Y / Wu H | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Mol Cell / Year: 2016 Journal: Mol Cell / Year: 2016Title: Cryo-EM Structure of Caspase-8 Tandem DED Filament Reveals Assembly and Regulation Mechanisms of the Death-Inducing Signaling Complex. Authors: Tian-Min Fu / Yang Li / Alvin Lu / Zongli Li / Parimala R Vajjhala / Anthony C Cruz / Devendra B Srivastava / Frank DiMaio / Pawel A Penczek / Richard M Siegel / Katryn J Stacey / Edward H Egelman / Hao Wu /   Abstract: Caspase-8 activation can be triggered by death receptor-mediated formation of the death-inducing signaling complex (DISC) and by the inflammasome adaptor ASC. Caspase-8 assembles with FADD at the ...Caspase-8 activation can be triggered by death receptor-mediated formation of the death-inducing signaling complex (DISC) and by the inflammasome adaptor ASC. Caspase-8 assembles with FADD at the DISC and with ASC at the inflammasome through its tandem death effector domain (tDED), which is regulated by the tDED-containing cellular inhibitor cFLIP and the viral inhibitor MC159. Here we present the caspase-8 tDED filament structure determined by cryoelectron microscopy. Extensive assembly interfaces not predicted by the previously proposed linear DED chain model were uncovered, and were further confirmed by structure-based mutagenesis in filament formation in vitro and Fas-induced apoptosis and ASC-mediated caspase-8 recruitment in cells. Structurally, the two DEDs in caspase-8 use quasi-equivalent contacts to enable assembly. Using the tDED filament structure as a template, structural analyses reveal the interaction surfaces between FADD and caspase-8 and the distinct mechanisms of regulation by cFLIP and MC159 through comingling and capping, respectively. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_8300.map.gz emd_8300.map.gz | 5.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-8300-v30.xml emd-8300-v30.xml emd-8300.xml emd-8300.xml | 10.2 KB 10.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_8300.png emd_8300.png | 126.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-8300 http://ftp.pdbj.org/pub/emdb/structures/EMD-8300 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8300 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-8300 | HTTPS FTP |
-Validation report
| Summary document |  emd_8300_validation.pdf.gz emd_8300_validation.pdf.gz | 528.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_8300_full_validation.pdf.gz emd_8300_full_validation.pdf.gz | 528.2 KB | Display | |
| Data in XML |  emd_8300_validation.xml.gz emd_8300_validation.xml.gz | 4.9 KB | Display | |
| Data in CIF |  emd_8300_validation.cif.gz emd_8300_validation.cif.gz | 5.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8300 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8300 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8300 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-8300 | HTTPS FTP |
-Related structure data
| Related structure data |  5l08MC  5jqeC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_8300.map.gz / Format: CCP4 / Size: 19.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_8300.map.gz / Format: CCP4 / Size: 19.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Casp-8 tDED filament | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
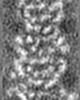
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.62 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
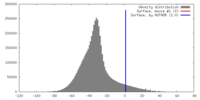
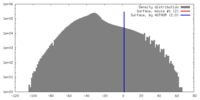
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Caspase-8
| Entire | Name: Caspase-8 |
|---|---|
| Components |
|
-Supramolecule #1: Caspase-8
| Supramolecule | Name: Caspase-8 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Caspase-8
| Macromolecule | Name: Caspase-8 / type: protein_or_peptide / ID: 1 / Enantiomer: LEVO / EC number: caspase-8 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  |
| Sequence | String: MDFSRNLYDI GEQLDSEDL A SLKFLSLD YI PQRKQEP IKD ALMLFQ RLQEKRMLEE SNLS FLKEL LFRIN RLDL LITYLNTRKE EMEREL QTP GRAQISAYRV MLYQISE EV SRSELRSF K FLLQEEISK CKLDDDMNLL DIFIEMEKR V ILGEGKLD IL ...String: MDFSRNLYDI GEQLDSEDL A SLKFLSLD YI PQRKQEP IKD ALMLFQ RLQEKRMLEE SNLS FLKEL LFRIN RLDL LITYLNTRKE EMEREL QTP GRAQISAYRV MLYQISE EV SRSELRSF K FLLQEEISK CKLDDDMNLL DIFIEMEKR V ILGEGKLD IL KRVCAQI NKS LLKIIN DYEE FSKE UniProtKB: Caspase-8 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Concentration | 0.2 mg/mL | ||||||
|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 / Component:
| ||||||
| Vitrification | Cryogen name: OTHER / Details: cryogen not identified. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI F20 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 31.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Sample stage | Specimen holder model: GATAN CT3500 SINGLE TILT LIQUID NITROGEN CRYO TRANSFER HOLDER Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Tecnai F20 / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 27.1 Å Applied symmetry - Helical parameters - Δ&Phi: 99.4 ° Applied symmetry - Helical parameters - Axial symmetry: C1 (asymmetric) Resolution.type: BY AUTHOR / Resolution: 4.6 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 33969 |
|---|---|
| Startup model | Type of model: INSILICO MODEL |
| Final angle assignment | Type: NOT APPLICABLE |
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-5l08: |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)