+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7476 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cdc48-Npl4 complex in the presence of ATP-gamma-S | |||||||||
 Map data Map data | primary map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | AAA+ / ERAD / zinc finger / ubiquitin-binding protein / MOTOR PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationnuclear protein quality control by the ubiquitin-proteasome system / mitotic spindle disassembly / VCP-NPL4-UFD1 AAA ATPase complex / retrograde protein transport, ER to cytosol / polyubiquitin modification-dependent protein binding / autophagosome maturation / mRNA transport / ubiquitin binding / protein transport / nuclear membrane ...nuclear protein quality control by the ubiquitin-proteasome system / mitotic spindle disassembly / VCP-NPL4-UFD1 AAA ATPase complex / retrograde protein transport, ER to cytosol / polyubiquitin modification-dependent protein binding / autophagosome maturation / mRNA transport / ubiquitin binding / protein transport / nuclear membrane / ubiquitin-dependent protein catabolic process / mitochondrial outer membrane / cell division / ubiquitin protein ligase binding / perinuclear region of cytoplasm / ATP hydrolysis activity / ATP binding / metal ion binding / nucleus / cytosol Similarity search - Function | |||||||||
| Biological species |  Chaetomium thermophilum (fungus) / Chaetomium thermophilum (fungus) /  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) | |||||||||
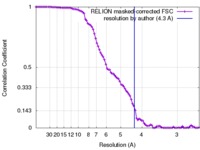
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Kim KH / Bodnar NO | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2018 Journal: Nat Struct Mol Biol / Year: 2018Title: Structure of the Cdc48 ATPase with its ubiquitin-binding cofactor Ufd1-Npl4. Authors: Nicholas O Bodnar / Kelly H Kim / Zhejian Ji / Thomas E Wales / Vladimir Svetlov / Evgeny Nudler / John R Engen / Thomas Walz / Tom A Rapoport /  Abstract: Many polyubiquitinated proteins are extracted from membranes or complexes by the conserved ATPase Cdc48 (in yeast; p97 or VCP in mammals) before proteasomal degradation. Each Cdc48 hexamer contains ...Many polyubiquitinated proteins are extracted from membranes or complexes by the conserved ATPase Cdc48 (in yeast; p97 or VCP in mammals) before proteasomal degradation. Each Cdc48 hexamer contains two stacked ATPase rings (D1 and D2) and six N-terminal (N) domains. Cdc48 binds various cofactors, including the Ufd1-Npl4 heterodimer. Here, we report structures of the Cdc48-Ufd1-Npl4 complex from Chaetomium thermophilum. Npl4 interacts through its UBX-like domain with a Cdc48 N domain, and it uses two Zn-finger domains to anchor the enzymatically inactive Mpr1-Pad1 N-terminal (MPN) domain, homologous to domains found in several isopeptidases, to the top of the D1 ATPase ring. The MPN domain of Npl4 is located above Cdc48's central pore, a position similar to the MPN domain from deubiquitinase Rpn11 in the proteasome. Our results indicate that Npl4 is unique among Cdc48 cofactors and suggest a mechanism for binding and translocation of polyubiquitinated substrates into the ATPase. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7476.map.gz emd_7476.map.gz | 5.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7476-v30.xml emd-7476-v30.xml emd-7476.xml emd-7476.xml | 15.9 KB 15.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7476_fsc.xml emd_7476_fsc.xml | 9 KB | Display |  FSC data file FSC data file |
| Images |  emd_7476.png emd_7476.png | 119 KB | ||
| Filedesc metadata |  emd-7476.cif.gz emd-7476.cif.gz | 6.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7476 http://ftp.pdbj.org/pub/emdb/structures/EMD-7476 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7476 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7476 | HTTPS FTP |
-Related structure data
| Related structure data |  6chsMC  7477C  7478C  7479C  6cddC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7476.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7476.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | primary map | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
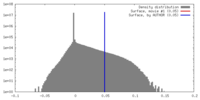
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cdc48-Npl4/Ufd1 complex
| Entire | Name: Cdc48-Npl4/Ufd1 complex |
|---|---|
| Components |
|
-Supramolecule #1: Cdc48-Npl4/Ufd1 complex
| Supramolecule | Name: Cdc48-Npl4/Ufd1 complex / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (fungus) Chaetomium thermophilum (fungus) |
-Macromolecule #1: Npl4
| Macromolecule | Name: Npl4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus)Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 73.880391 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLLRMRCPDG MFRLTVEKDD TFGELVRQLV PKLPPTVDPK SITLSNHPSG GDSKRIDEIA RFKIGQVGHG DLIFVRYQTS DTVANGRSV DISSQTAGLS SSANRLNGKP VLPTEDHPID PPPNTSAERI KNPWEVVRQS PLDDRLDKLD GKIPRKRGAM C RHGPKGMC ...String: MLLRMRCPDG MFRLTVEKDD TFGELVRQLV PKLPPTVDPK SITLSNHPSG GDSKRIDEIA RFKIGQVGHG DLIFVRYQTS DTVANGRSV DISSQTAGLS SSANRLNGKP VLPTEDHPID PPPNTSAERI KNPWEVVRQS PLDDRLDKLD GKIPRKRGAM C RHGPKGMC DYCTPLDPFN PQYLEEKKIK YMSVHAYMRK INSATNRPEL GSSFIPPLVE PYYRVKRDCP SGHPQWPEGI CT KCQPSAI TLQPQPFRMV DHVEFASPQI IDRFLDAWRR TGVQRLGILY GRYLEYDAVP LGIKAVVEAI YEPPQVDEID GIT LNPWEN EQEVNQVAKY CGLEQVGVIW TDLLDAGKGD GSVVCKRHAD SYFLAAQEIV FAARLQAQHP KPSKWSDTGR FGSN FVTCV VSGNEQGEIS ISAYQMSNDA VEMVRADIIE PSADPTLMLV REEEEDDGSP SKTRYIPEVF YRKINEYGAN VLENA KPAF PVEYLFVTLT HGFPDSPSPL FTDNIFPIEN REYVGEAQEV SAVAKALKVH EADAPMNVSD FHLLCFIHQM SVLSKE EEA LLCRVATLHD LAESFQLRST TGWQTLHMIL QSTGERVPKR PRFSDNVPSS SQDTQEALGG NNLDPGEPLA KRFAAVR LN ENNQSSPKPY APSRWNL UniProtKB: Nuclear protein localization protein 4 |
-Macromolecule #2: Putative cell division control protein
| Macromolecule | Name: Putative cell division control protein / type: protein_or_peptide / ID: 2 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus) Chaetomium thermophilum (strain DSM 1495 / CBS 144.50 / IMI 039719) (fungus)Strain: DSM 1495 / CBS 144.50 / IMI 039719 |
| Molecular weight | Theoretical: 90.432516 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAPESKDHKK KVNLTDPSGA EHREENDTAT AILKKKKKPN QLMVTDAVND DNSIIALSNN TMEALQLFRG DTVLVRGKKR RDTVLIVLA DDDLDDGSAR INRVVRHNLR VKHGDMITIH PCPDIKYAKR IAVLPIADTV EGLTGSLFDV FLAPYFREAY R PVRQGDLF ...String: MAPESKDHKK KVNLTDPSGA EHREENDTAT AILKKKKKPN QLMVTDAVND DNSIIALSNN TMEALQLFRG DTVLVRGKKR RDTVLIVLA DDDLDDGSAR INRVVRHNLR VKHGDMITIH PCPDIKYAKR IAVLPIADTV EGLTGSLFDV FLAPYFREAY R PVRQGDLF TVRGGMRQVE FKVVEVDPPE YGIVAQDTII HCEGEPIPRE EEENNLNEVG YDDIGGCRKQ LAQIREMVEL PL RHPQLFK SIGIKPPRGV LLYGPPGTGK TLMARAVANE TGAFFFLING PEIMSKMAGE SESNLRKAFE EAEKNSPAII FID EIDSIA PKREKTNGEV ERRVVSQLLT LMDGMKARSN VVVMAATNRP NSIDPALRRF GRFDREVDIG IPDPTGRLEI LQIH TKNMK LADDVDLEQI AAETHGYVGS DLAALCSEAA MQQIREKMDL IDLDEDTIDA EVLDSLGVTM DNFRYALGVS NPSAL REVA VVEVPNVRWE DIGGLEQVKQ ELKEQVQYPV DHPEKFLKFG LSPSRGVLFY GPPGTGKTML AKAVANECAA NFISVK GPE LLSMWFGESE SNIRDIFDKA RAAAPCVVFL DELDSIAKAR GGSIGDAGGA SDRVVNQLLT EMDGMTSKKN VFVIGAT NR PEQLDPALCR PGRLDQLIYV PLPDEAGRLS ILKAQLRKTP VSKDVDLAYI ASKTHGFSGA DLAFITQRAV KLAIKESI A AEIERQKARE AAGEDVNMED DEDPVPELTK RHFEEAMRDA RRSVSDVEIR RYEAFAQQMK NAGPGAFFKF PDSTTDNSA SNAAGNSFGD AGNDDDLYT UniProtKB: Putative cell division control protein |
-Macromolecule #3: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 3 / Number of copies: 2 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #4: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER / type: ligand / ID: 4 / Number of copies: 12 / Formula: AGS |
|---|---|
| Molecular weight | Theoretical: 523.247 Da |
| Chemical component information |  ChemComp-AGS: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 12 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| ||||||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 80 % / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 9299 / Average exposure time: 15.0 sec. / Average electron dose: 1.8 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT |
|---|---|
| Output model |  PDB-6chs: |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)