[English] 日本語
 Yorodumi
Yorodumi- EMDB-7344: An anti-gH/gL antibody that neutralizes dual-tropic infection def... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7344 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
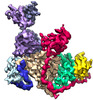
| Title | An anti-gH/gL antibody that neutralizes dual-tropic infection defines a site of vulnerability on Epstein-Barr virus | |||||||||
 Map data Map data | An anti-gH/gL antibody that neutralizes dual-tropic infection defines a site of vulnerability on Epstein-Barr virus | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Epstein-Barr virus / neutralizing antibodies / gH/gL / glycoproteins / Structural Genomics / Seattle Structural Genomics Center for Infectious Disease / SSGCID / VIRAL PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationmembrane => GO:0016020 / host cell endosome membrane / carbohydrate binding / host cell Golgi apparatus / fusion of virus membrane with host plasma membrane / viral envelope / symbiont entry into host cell / host cell plasma membrane / virion membrane / membrane Similarity search - Function | |||||||||
| Biological species |  Human herpesvirus 4 (Epstein-Barr virus) / Human herpesvirus 4 (Epstein-Barr virus) /  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.8 Å | |||||||||
 Authors Authors | Snijder J / Ortego MS | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Immunity / Year: 2018 Journal: Immunity / Year: 2018Title: An Antibody Targeting the Fusion Machinery Neutralizes Dual-Tropic Infection and Defines a Site of Vulnerability on Epstein-Barr Virus. Authors: Joost Snijder / Michael S Ortego / Connor Weidle / Andrew B Stuart / Matthew D Gray / M Juliana McElrath / Marie Pancera / David Veesler / Andrew T McGuire /  Abstract: Epstein-Barr virus (EBV) is a causative agent of infectious mononucleosis and is associated with 200,000 new cases of cancer and 140,000 deaths annually. Subunit vaccines against this pathogen have ...Epstein-Barr virus (EBV) is a causative agent of infectious mononucleosis and is associated with 200,000 new cases of cancer and 140,000 deaths annually. Subunit vaccines against this pathogen have focused on the gp350 glycoprotein and remain unsuccessful. We isolated human antibodies recognizing the EBV fusion machinery (gH/gL and gB) from rare memory B cells. One anti-gH/gL antibody, AMMO1, potently neutralized infection of B cells and epithelial cells, the two major cell types targeted by EBV. We determined a cryo-electron microscopy reconstruction of the gH/gL-gp42-AMMO1 complex and demonstrated that AMMO1 bound to a discontinuous epitope formed by both gH and gL at the Domain-I/Domain-II interface. Integrating structural, biochemical, and infectivity data, we propose that AMMO1 inhibits fusion of the viral and cellular membranes. This work identifies a crucial epitope that may aid in the design of next-generation subunit vaccines against this major public health burden. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7344.map.gz emd_7344.map.gz | 3.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7344-v30.xml emd-7344-v30.xml emd-7344.xml emd-7344.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7344.png emd_7344.png | 205.6 KB | ||
| Filedesc metadata |  emd-7344.cif.gz emd-7344.cif.gz | 6.6 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7344 http://ftp.pdbj.org/pub/emdb/structures/EMD-7344 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7344 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7344 | HTTPS FTP |
-Validation report
| Summary document |  emd_7344_validation.pdf.gz emd_7344_validation.pdf.gz | 346.7 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7344_full_validation.pdf.gz emd_7344_full_validation.pdf.gz | 346.3 KB | Display | |
| Data in XML |  emd_7344_validation.xml.gz emd_7344_validation.xml.gz | 5.9 KB | Display | |
| Data in CIF |  emd_7344_validation.cif.gz emd_7344_validation.cif.gz | 6.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7344 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7344 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7344 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7344 | HTTPS FTP |
-Related structure data
| Related structure data |  6c5vMC  7345C  6blaC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7344.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7344.map.gz / Format: CCP4 / Size: 42.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | An anti-gH/gL antibody that neutralizes dual-tropic infection defines a site of vulnerability on Epstein-Barr virus | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.36 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Epstein-Barr virus glycoproteins H,L,42 in complex with the human...
| Entire | Name: Epstein-Barr virus glycoproteins H,L,42 in complex with the human antibody Fab fragment AMMO1. |
|---|---|
| Components |
|
-Supramolecule #1: Epstein-Barr virus glycoproteins H,L,42 in complex with the human...
| Supramolecule | Name: Epstein-Barr virus glycoproteins H,L,42 in complex with the human antibody Fab fragment AMMO1. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|
-Supramolecule #2: Epstein-Barr virus
| Supramolecule | Name: Epstein-Barr virus / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Human herpesvirus 4 (Epstein-Barr virus) Human herpesvirus 4 (Epstein-Barr virus) |
-Supramolecule #3: Human
| Supramolecule | Name: Human / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #4-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Envelope glycoprotein H
| Macromolecule | Name: Envelope glycoprotein H / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human herpesvirus 4 (Epstein-Barr virus) / Strain: GD1 Human herpesvirus 4 (Epstein-Barr virus) / Strain: GD1 |
| Molecular weight | Theoretical: 77.091547 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDAMKRGLCC VLLLCGAVFV SPSASSLSEV KLHLDIEGHA SHYTIPWTEL MAKVPGLSPE ALWREANVTE DLASMLNRYK LIYKTSGTL GIALAEPVDI PAVSEGSMQV DASKVHPGVI SGLNSPACML SAPLEKQLFY YIGTMLPNTR PHSYVFYQLR C HLSYVALS ...String: MDAMKRGLCC VLLLCGAVFV SPSASSLSEV KLHLDIEGHA SHYTIPWTEL MAKVPGLSPE ALWREANVTE DLASMLNRYK LIYKTSGTL GIALAEPVDI PAVSEGSMQV DASKVHPGVI SGLNSPACML SAPLEKQLFY YIGTMLPNTR PHSYVFYQLR C HLSYVALS INGDKFQYTG AMTSKFLMGT YKRVTEKGDE HVLSLVFGKT KDLPDLRGPF SYPSLTSAQS GDYSLVIVTT FV HYANFHN YFVPNLKDMF SRAVTMTAAS YARYVLQKLV LLEMKGGCRE PELDTETLTT MFEVSVAFFK VGHAVGETGN GCV DLRWLA KSFFELTVLK DIIGICYGAT VKGMQSYGLE RLAAMLMATV KMEELGHLTT EKQEYALRLA TVGYPKAGVY SGLI GGATS VLLSAYNRHP LFQPLHTVMR ETLFIGSHVV LRELRLNVTT QGPNLALYQL LSTALCSALE IGEVLRGLAL GTESG LFSP CYLSLRFDLT RDKLLSMAPQ EATLDQAAVS YAVDGFLGRL SLEREDRDAW HLPAYKCVDR LDKVLMIIPL INVTFI ISS DREVRGSALY EASTTYLSSS LFLSPVIMNK CSQGAVAGEP RQIPKIQNFT RTQKSCIFCG FALLSYDEKE GLETTTY IT SQEVQNSILS SNYFDFDNLH VHYLLLTTNG TVMEIAGLYE ERAHGSGSGH HHHHH UniProtKB: Envelope glycoprotein H |
-Macromolecule #2: Envelope glycoprotein L
| Macromolecule | Name: Envelope glycoprotein L / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human herpesvirus 4 (Epstein-Barr virus) / Strain: AG876 Human herpesvirus 4 (Epstein-Barr virus) / Strain: AG876 |
| Molecular weight | Theoretical: 15.145395 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDAMKRGLCC VLLLCGAVFV SPSASWAYPC CHVTQLRAQH LLALENISDI YLVSNQTCDG FSLASLNSPK NGSNQLVISR CANGLNVVS FFISILKRSS SALTGHLREL LTTLETLYGS FSVEDLFGAN LNRYAWHRGG UniProtKB: Envelope glycoprotein L |
-Macromolecule #3: Glycoprotein 42
| Macromolecule | Name: Glycoprotein 42 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Human herpesvirus 4 (Epstein-Barr virus) / Strain: GD1 Human herpesvirus 4 (Epstein-Barr virus) / Strain: GD1 |
| Molecular weight | Theoretical: 25.356859 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDAMKRGLCC VLLLCGAVFV SPSASGGGRV AAAAITWVPK PNVEVWPVDP PPPVNFNKTA EQEYGDKEVK LPHWTPTLHT FQVPQNYTK ANCTYCNTRE YTFSYKGCCF YFTKKKHTWN GCFQACAELY PCTYFYGPTP DILPVVTRNL NAIESLWVGV Y RVGEGNWT ...String: MDAMKRGLCC VLLLCGAVFV SPSASGGGRV AAAAITWVPK PNVEVWPVDP PPPVNFNKTA EQEYGDKEVK LPHWTPTLHT FQVPQNYTK ANCTYCNTRE YTFSYKGCCF YFTKKKHTWN GCFQACAELY PCTYFYGPTP DILPVVTRNL NAIESLWVGV Y RVGEGNWT SLDGGTFKVY QIFGSHCTYV SKFSTVPVSH HECSFLKPCL CVSQRSNSGS GSGHHHHHH UniProtKB: Glycoprotein 42 |
-Macromolecule #4: Antibody Fab AMMO1 heavy chain
| Macromolecule | Name: Antibody Fab AMMO1 heavy chain / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.982885 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: (PCA)VQLVQSGAD VKKPGASVKV SCKASGYTFI HFGISWVRQA PGQGLEWMGW IDTNNGNTNY AQSLQGRVTM TTDTST GTA YMELRSLSTD DTAVYFCARA LEMGHRSGFP FDYWGQGVLV TVSPASTKGP SVFPLAPSSK STSGGTAALG CLVKDYF PE PVTVSWNSGA ...String: (PCA)VQLVQSGAD VKKPGASVKV SCKASGYTFI HFGISWVRQA PGQGLEWMGW IDTNNGNTNY AQSLQGRVTM TTDTST GTA YMELRSLSTD DTAVYFCARA LEMGHRSGFP FDYWGQGVLV TVSPASTKGP SVFPLAPSSK STSGGTAALG CLVKDYF PE PVTVSWNSGA LTSGVHTFPA VLQSSGLYSL SSVVTVPSSS LGTQTYICNV NHKPSNTKVD KRVEPKSC |
-Macromolecule #5: Antibody Fab AMMO1 light chain
| Macromolecule | Name: Antibody Fab AMMO1 light chain / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.095514 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SYELTQPPSV SVAPGQRATI TCGGHNIGAK NVHWYQQKPG QAPVLVIQYD SDRPSGIPER FSGSNSGSTA TLTISRVEAG DEADYYCQV WDSGRGHPLY VFGGGTKVTV LGQPKANPTV TLFPPSSEEL QANKATLVCL ISDFYPGAVT VAWKADSSPV K AGVETTTP ...String: SYELTQPPSV SVAPGQRATI TCGGHNIGAK NVHWYQQKPG QAPVLVIQYD SDRPSGIPER FSGSNSGSTA TLTISRVEAG DEADYYCQV WDSGRGHPLY VFGGGTKVTV LGQPKANPTV TLFPPSSEEL QANKATLVCL ISDFYPGAVT VAWKADSSPV K AGVETTTP SKQSNNKYAA SSYLSLTPEQ WKSHRSYSCQ VTHEGSTVEK TVAPTECS |
-Macromolecule #7: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 7 / Number of copies: 8 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 65.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 4.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 72000 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















