[English] 日本語
 Yorodumi
Yorodumi- EMDB-7076: Mechanisms of Opening and Closing of the Bacterial Replicative He... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7076 | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Mechanisms of Opening and Closing of the Bacterial Replicative Helicase: The DnaB Helicase and Lambda P Helicase Loader Complex | |||||||||||||||||||||||||||||||||||||||
 Map data Map data | DnaB-LambdaP helicase-helicase loader complex from single particle cryoEM at 4.1A. The suggested viewing thresholds are 0.0276 (Chimera) or an isomesh level of 6 (PyMol). | |||||||||||||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||||||||||||||
 Keywords Keywords | Helicase Loader / Helicase / DNA replication / ATPase / DNA Replication Initiation / Bacteriophage Lambda / REPLICATION | |||||||||||||||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationDnaB-DnaC complex / DnaB-DnaC-Rep-PriC complex / DnaB-DnaG complex / DnaB-DnaC-DnaT-PriA-PriC complex / DnaB-DnaC-DnaT-PriA-PriB complex / DNA helicase complex / primosome complex / DNA 5'-3' helicase / bidirectional double-stranded viral DNA replication / DNA replication, synthesis of primer ...DnaB-DnaC complex / DnaB-DnaC-Rep-PriC complex / DnaB-DnaG complex / DnaB-DnaC-DnaT-PriA-PriC complex / DnaB-DnaC-DnaT-PriA-PriB complex / DNA helicase complex / primosome complex / DNA 5'-3' helicase / bidirectional double-stranded viral DNA replication / DNA replication, synthesis of primer / replisome / response to ionizing radiation / replication fork processing / DNA replication initiation / DNA helicase activity / helicase activity / 5'-3' DNA helicase activity / DNA helicase / DNA replication / hydrolase activity / ATP hydrolysis activity / DNA binding / ATP binding / identical protein binding / cytosol Similarity search - Function | |||||||||||||||||||||||||||||||||||||||
| Biological species |   Enterobacteria phage lambda (virus) / Enterobacteria phage lambda (virus) /   Escherichia phage lambda (virus) Escherichia phage lambda (virus) | |||||||||||||||||||||||||||||||||||||||
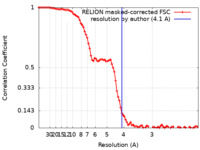
| Method | single particle reconstruction / cryo EM / Resolution: 4.1 Å | |||||||||||||||||||||||||||||||||||||||
 Authors Authors | Chase J / Catalano A | |||||||||||||||||||||||||||||||||||||||
| Funding support |  United States, 12 items United States, 12 items
| |||||||||||||||||||||||||||||||||||||||
 Citation Citation |  Journal: Elife / Year: 2018 Journal: Elife / Year: 2018Title: Mechanisms of opening and closing of the bacterial replicative helicase. Authors: Jillian Chase / Andrew Catalano / Alex J Noble / Edward T Eng / Paul Db Olinares / Kelly Molloy / Danaya Pakotiprapha / Martin Samuels / Brian Chait / Amedee des Georges / David Jeruzalmi /   Abstract: Assembly of bacterial ring-shaped hexameric replicative helicases on single-stranded (ss) DNA requires specialized loading factors. However, mechanisms implemented by these factors during opening and ...Assembly of bacterial ring-shaped hexameric replicative helicases on single-stranded (ss) DNA requires specialized loading factors. However, mechanisms implemented by these factors during opening and closing of the helicase, which enable and restrict access to an internal chamber, are not known. Here, we investigate these mechanisms in the DnaB helicase•bacteriophage λ helicase loader (λP) complex. We show that five copies of λP bind at DnaB subunit interfaces and reconfigure the helicase into an open spiral conformation that is intermediate to previously observed closed ring and closed spiral forms; reconfiguration also produces openings large enough to admit ssDNA into the inner chamber. The helicase is also observed in a restrained inactive configuration that poises it to close on activating signal, and transition to the translocation state. Our findings provide insights into helicase opening, delivery to the origin and ssDNA entry, and closing in preparation for translocation. | |||||||||||||||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7076.map.gz emd_7076.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7076-v30.xml emd-7076-v30.xml emd-7076.xml emd-7076.xml | 26.1 KB 26.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_7076_fsc.xml emd_7076_fsc.xml | 9.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_7076.png emd_7076.png | 157.3 KB | ||
| Filedesc metadata |  emd-7076.cif.gz emd-7076.cif.gz | 9.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7076 http://ftp.pdbj.org/pub/emdb/structures/EMD-7076 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7076 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7076 | HTTPS FTP |
-Validation report
| Summary document |  emd_7076_validation.pdf.gz emd_7076_validation.pdf.gz | 611.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_7076_full_validation.pdf.gz emd_7076_full_validation.pdf.gz | 610.7 KB | Display | |
| Data in XML |  emd_7076_validation.xml.gz emd_7076_validation.xml.gz | 10.7 KB | Display | |
| Data in CIF |  emd_7076_validation.cif.gz emd_7076_validation.cif.gz | 14.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7076 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7076 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7076 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-7076 | HTTPS FTP |
-Related structure data
| Related structure data |  6bbmMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_7076.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7076.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | DnaB-LambdaP helicase-helicase loader complex from single particle cryoEM at 4.1A. The suggested viewing thresholds are 0.0276 (Chimera) or an isomesh level of 6 (PyMol). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : DnaB helicase - Lambda P helicase loader DNA replication complex
| Entire | Name: DnaB helicase - Lambda P helicase loader DNA replication complex |
|---|---|
| Components |
|
-Supramolecule #1: DnaB helicase - Lambda P helicase loader DNA replication complex
| Supramolecule | Name: DnaB helicase - Lambda P helicase loader DNA replication complex type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 Details: pET24a containing full-length DnaB was co-expressed with pCDFDuet containing full-length LambdaP in BL21(DE3) cells. The resolution of the LambdaP portion of our EM map did not permit the ...Details: pET24a containing full-length DnaB was co-expressed with pCDFDuet containing full-length LambdaP in BL21(DE3) cells. The resolution of the LambdaP portion of our EM map did not permit the unambiguous assignment of the amino acid sequence to the structure. As such, the model for LambdaP was built as a poly alanine model. Additionally, only half of LambdaP was observed in our maps due to the intrinsic flexibility of the amino and carboxy terminal domains of LambdaP. Subsequent experiments determined that the observed portion of LambdaP in our maps corresponds to the C-terminal domain. |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: E coli DnaB helicase
| Supramolecule | Name: E coli DnaB helicase / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 Details: E coli DnaB helicase is observed as an open-spiral hexamer, in which one of the interfaces is breached. Five ADP molecules are observed at the five intact ATP binding sites. Additionally, ...Details: E coli DnaB helicase is observed as an open-spiral hexamer, in which one of the interfaces is breached. Five ADP molecules are observed at the five intact ATP binding sites. Additionally, clear density is observed for five of six linkers permitting unambiguous assignment of NTD to parent CTD domain. |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: Lambda P helicase loader
| Supramolecule | Name: Lambda P helicase loader / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 Details: Five lambda P molecules were observed bound to the five intact DnaB subunit interfaces. Unambiguous assignment of side chain density for lambda P was not possible due to the resolution of ...Details: Five lambda P molecules were observed bound to the five intact DnaB subunit interfaces. Unambiguous assignment of side chain density for lambda P was not possible due to the resolution of this region of the EM map. Instead, a polyalanine model was built for each lambda P molecule. Additionally, density for approximately half of the expected 233 residues of lambda P was observed owing to flexibility between domains. Subsequent experiments confirmed that the observed region of Lambda P is the C-terminal domain, which interacts with DnaB. |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage lambda (virus) Enterobacteria phage lambda (virus) |
-Macromolecule #1: Replicative DNA helicase
| Macromolecule | Name: Replicative DNA helicase / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO / EC number: DNA helicase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 52.450945 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAGNKPFNKQ QAEPRERDPQ VAGLKVPPHS IEAEQSVLGG LMLDNERWDD VAERVVADDF YTRPHRHIFT EMARLQESGS PIDLITLAE SLERQGQLDS VGGFAYLAEL SKNTPSAANI SAYADIVRER AVVREMISVA NEIAEAGFDP QGRTSEDLLD L AESRVFKI ...String: MAGNKPFNKQ QAEPRERDPQ VAGLKVPPHS IEAEQSVLGG LMLDNERWDD VAERVVADDF YTRPHRHIFT EMARLQESGS PIDLITLAE SLERQGQLDS VGGFAYLAEL SKNTPSAANI SAYADIVRER AVVREMISVA NEIAEAGFDP QGRTSEDLLD L AESRVFKI AESRANKDEG PKNIADVLDA TVARIEQLFQ QPHDGVTGVN TGYDDLNKKT AGLQPSDLII VAARPSMGKT TF AMNLVEN AAMLQDKPVL IFSLEMPSEQ IMMRSLASLS RVDQTKIRTG QLDDEDWARI SGTMGILLEK RNIYIDDSSG LTP TEVRSR ARRIAREHGG IGLIMIDYLQ LMRVPALSDN RTLEIAEISR SLKALAKELN VPVVALSQLN RSLEQRADKR PVNS DLRES GSIEQDADLI MFIYRDEVYH ENSDLKGIAE IIIGKQRNGP IGTVRLTFNG QWSRFDNYAG PQYDDE UniProtKB: Replicative DNA helicase |
-Macromolecule #2: Replication protein P
| Macromolecule | Name: Replication protein P / type: protein_or_peptide / ID: 2 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Escherichia phage lambda (virus) Escherichia phage lambda (virus) |
| Molecular weight | Theoretical: 23.141221 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKNIAAQMVN FDREQMRRIA NNMPEQYDEK PQVQQVAQII NGVFSQLLAT FPASLANRDQ NEVNEIRRQW VLAFRENGIT TMEQVNAGM RVARRQNRPF LPSPGQFV(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK) ...String: MKNIAAQMVN FDREQMRRIA NNMPEQYDEK PQVQQVAQII NGVFSQLLAT FPASLANRDQ NEVNEIRRQW VLAFRENGIT TMEQVNAGM RVARRQNRPF LPSPGQFV(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK) UniProtKB: Helicase loader |
-Macromolecule #3: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 3 / Number of copies: 5 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.48 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
Details: Concentrated BP sample (18mg/mL) was diluted with freshly prepared buffer to desired concentration (~1.5 micromolar) for grid preparation. | ||||||||||||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: GOLD / Mesh: 400 / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER Details: The grid was coated with 50 nm of evaporated gold prior to use. All remaining carbon was removed by plasma cleaning for 5 minutes in a Gatan Solarus plasma cleaner. | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV Details: 3uL of sample was adhered to a fresh plasma cleaned grid and allowed to adsorb for 30 seconds, blotted for 3 seconds with a blot force of 4 and plunge frozen into liquid nitrogen-cooled ethane.. | ||||||||||||||||||
| Details | The sample was monodisperse. Side views were more electron weak than top or bottom views creating challenges for particle picking. This issue was overcome with cryo-electron tomography techniques used for 1) initial model generation and 2) template generation for particle picking. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 70.0 K / Max: 70.0 K |
| Specialist optics | Spherical aberration corrector: The Krios this data was collected on has a Cs of 2.7. |
| Details | Preliminary grid screening was performed prior to Krios data collections. All microscope alignments were completed by the New York Structural Biology SEMC team. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Dimensions - Width: 3838 pixel / Digitization - Dimensions - Height: 3710 pixel / Digitization - Frames/image: 1-50 / Number grids imaged: 3 / Number real images: 2426 / Average exposure time: 10.0 sec. / Average electron dose: 8.0 e/Å2 Details: Single particle movies were recorded at a pixel size of 1.07 angstroms/pixel. Three 24-hour sessions produced 2,426 micrograph movies. In addition, five tilt series were collected from the ...Details: Single particle movies were recorded at a pixel size of 1.07 angstroms/pixel. Three 24-hour sessions produced 2,426 micrograph movies. In addition, five tilt series were collected from the same grids bi-directionally over a tilt range of -45 degrees to +45 degrees in 3 degree increments at a dose of 2.57 to 3.3 electrons per angstrom squared (total accumulated dose of 90 electrons per angstrom squared). Tilt series were collected at a pixel size of 1.76 angstroms and at defocus values of -2.8um, -6.1um and -9.3um. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: -0.003 µm / Nominal defocus min: -0.001 µm / Nominal magnification: 22500 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Details | The initial fitting was done with the 2R5U and 3BH0 models onto which the E. coli amino sequence had been built. The linker segments that connected these segments were built by hand. PHENIX real_space_refine was used to refine the complete model for the B6P5 entity. | ||||||
| Refinement | Space: REAL / Protocol: FLEXIBLE FIT | ||||||
| Output model |  PDB-6bbm: |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)