[English] 日本語
 Yorodumi
Yorodumi- EMDB-6963: Fit R10 Fab coordinates into the cryo-EM of EV71 in complex with D6 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6963 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Fit R10 Fab coordinates into the cryo-EM of EV71 in complex with D6 | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | EV71 NEUTRALIZING ANTIBODY / ANTIVIRAL PROTEIN / VIRUS LIKE PARTICLE | |||||||||
| Function / homology |  Function and homology information Function and homology informationsymbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / virion component / ribonucleoside triphosphate phosphatase activity / viral capsid ...symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of MDA-5 activity / picornain 2A / symbiont-mediated suppression of host mRNA export from nucleus / symbiont genome entry into host cell via pore formation in plasma membrane / picornain 3C / T=pseudo3 icosahedral viral capsid / host cell cytoplasmic vesicle membrane / virion component / ribonucleoside triphosphate phosphatase activity / viral capsid / nucleoside-triphosphate phosphatase / host cell / channel activity / monoatomic ion transmembrane transport / host cell cytoplasm / DNA replication / RNA helicase activity / endocytosis involved in viral entry into host cell / symbiont-mediated suppression of host gene expression / symbiont-mediated activation of host autophagy / RNA-directed RNA polymerase / cysteine-type endopeptidase activity / viral RNA genome replication / RNA-directed RNA polymerase activity / symbiont entry into host cell / DNA-templated transcription / virion attachment to host cell / host cell nucleus / structural molecule activity / proteolysis / RNA binding / zinc ion binding / ATP binding Similarity search - Function | |||||||||
| Biological species |   Enterovirus A71 / Enterovirus A71 /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.9 Å | |||||||||
 Authors Authors | Wang X / Zhu L | |||||||||
 Citation Citation |  Journal: mBio / Year: 2018 Journal: mBio / Year: 2018Title: Neutralization Mechanisms of Two Highly Potent Antibodies against Human Enterovirus 71. Authors: Ling Zhu / Kangwei Xu / Nan Wang / Lei Cao / Junlan Wu / Qiang Gao / Elizabeth E Fry / David I Stuart / Zihe Rao / Junzhi Wang / Xiangxi Wang /   Abstract: Despite significant advances in health care, outbreaks of infections by enteroviruses (EVs) continue to plague the Asia-Pacific region every year. Enterovirus 71 (EV71) causes hand-foot-and-mouth ...Despite significant advances in health care, outbreaks of infections by enteroviruses (EVs) continue to plague the Asia-Pacific region every year. Enterovirus 71 (EV71) causes hand-foot-and-mouth disease (HFMD), for which there are currently no therapeutics. Here, we report two new antibodies, A9 and D6, that potently neutralize EV71. A9 exhibited a 50% neutralizing concentration (neut) value of 0.1 nM against EV71, which was 10-fold lower than that observed for D6. Investigation into the mechanisms of neutralization revealed that binding of A9 to EV71 blocks receptor binding but also destabilizes and damages the virus capsid structure. In contrast, D6 destabilizes the capsid only slightly but interferes more potently with the attachment of the virus to the host cells. Cryo-electron microscopy (cryo-EM) structures of A9 and D6 bound with EV71 shed light on the locations and nature of the epitopes recognized by the two antibodies. Although some regions of the epitopes recognized by the two antibodies overlap, there are differences that give rise to dissimilarities in potency as well as in the mechanisms of neutralization. Interestingly, the overlapping regions of the epitopes encompass the site that the virus uses to bind SCARB2, explaining the reason for the observed blocking of the virus-receptor interaction by the two antibodies. We also identified structural elements that might play roles in modulating the stability of the EV71 particles, including particle integrity. The molecular features of the A9 and D6 epitopes unveiled in this study open up new avenues for rationally designing antiviral drugs. During the course of viral infections, the human body produces neutralizing antibodies which play a defining role in clearing the virus. From this study, we report two new, highly potent neutralizing antibodies, A9 and D6, against enterovirus 71 (EV71), the causative agent of HFMD. Both antibodies prevent the virus from entering the host cell, a step that is important for establishing a successful infection. A9 destabilizes and damages the virus capsid that forms an outer protective covering around the genome of the virus, while also interfering with virus attachment to the host cells. In contrast, D6 only prevents binding of the virus to its receptor(s). The mechanism of neutralization of A9 is unique and has not been observed before for neutralizing antibodies targeting EVs. The two antibodies that we are reporting in this study have potential to be developed into much-needed therapeutic interventions for treatment of HFMD, outbreaks of which are reported every year in the Asia-Pacific region. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6963.map.gz emd_6963.map.gz | 395.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6963-v30.xml emd-6963-v30.xml emd-6963.xml emd-6963.xml | 15 KB 15 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6963.png emd_6963.png | 195.4 KB | ||
| Filedesc metadata |  emd-6963.cif.gz emd-6963.cif.gz | 5.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6963 http://ftp.pdbj.org/pub/emdb/structures/EMD-6963 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6963 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6963 | HTTPS FTP |
-Related structure data
| Related structure data |  5zudMC  6964C  5zufC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6963.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6963.map.gz / Format: CCP4 / Size: 421.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
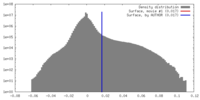
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : COMPLEX OF EV71 AND D6 NEUTRALIZING ANTIBODY FAB
| Entire | Name: COMPLEX OF EV71 AND D6 NEUTRALIZING ANTIBODY FAB |
|---|---|
| Components |
|
-Supramolecule #1: COMPLEX OF EV71 AND D6 NEUTRALIZING ANTIBODY FAB
| Supramolecule | Name: COMPLEX OF EV71 AND D6 NEUTRALIZING ANTIBODY FAB / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all Details: D6 FAB GENERATED BY MICE AND CLEAVAGE BY ENZYME FROM ANTIBODY |
|---|
-Supramolecule #2: EV71
| Supramolecule | Name: EV71 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#3 / Details: capsid protein VP1,VP2,VP3 |
|---|---|
| Source (natural) | Organism:   Enterovirus A71 Enterovirus A71 |
-Supramolecule #3: D6 NEUTRALIZING ANTIBODY FAB
| Supramolecule | Name: D6 NEUTRALIZING ANTIBODY FAB / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #4-#5 / Details: antibodies |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Capsid protein VP1
| Macromolecule | Name: Capsid protein VP1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Enterovirus A71 Enterovirus A71 |
| Molecular weight | Theoretical: 32.787902 KDa |
| Recombinant expression | Organism:  Chlorocebus aethiops (grivet monkey) Chlorocebus aethiops (grivet monkey) |
| Sequence | String: GDRVADVIES SIEDSSSRAL THALPAPTGQ NTQVSSHRLD TGKVPALQAA EIGASSNASD ESMIETRCVL NSHSTAETTL DSFFSRAGL VGEIDLPLKG TTNPNGYANW DIDITGYAQM RRKVELFTYM RFDAEFTFVA CTPTGEVVPQ LLQYMFVPPG A PKPDSRES ...String: GDRVADVIES SIEDSSSRAL THALPAPTGQ NTQVSSHRLD TGKVPALQAA EIGASSNASD ESMIETRCVL NSHSTAETTL DSFFSRAGL VGEIDLPLKG TTNPNGYANW DIDITGYAQM RRKVELFTYM RFDAEFTFVA CTPTGEVVPQ LLQYMFVPPG A PKPDSRES LAWQTATNPS VFVKLSDPPA QVSVPFMSPA SAYQWFYDGY PTFGEHKQEK DLEYGAMPNN MMGTFSVRTV GT SKSKYPL VVRIYMRMKH VRAWIPRPMR NQNYLFKANP NYAGNSIKPT GASRTAITTL UniProtKB: Genome polyprotein |
-Macromolecule #2: VP2
| Macromolecule | Name: VP2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Enterovirus A71 Enterovirus A71 |
| Molecular weight | Theoretical: 26.874252 KDa |
| Recombinant expression | Organism:  Chlorocebus aethiops (grivet monkey) Chlorocebus aethiops (grivet monkey) |
| Sequence | String: SDRVAQLTIG NSTITTQEAA NIIVGYGEWP SYCSDSDATA VDKPTRPDVS VNRFYTLDTK LWEKSSKGWY WKFPDVLTET GVFGQNAQF HYLYRSGFCI HVQCNASKFH QGALLVAVLP EYVIGTVAGG TGTEDTHPPY KQTQPGADGF ELQHPYVLDA G IPISQLTV ...String: SDRVAQLTIG NSTITTQEAA NIIVGYGEWP SYCSDSDATA VDKPTRPDVS VNRFYTLDTK LWEKSSKGWY WKFPDVLTET GVFGQNAQF HYLYRSGFCI HVQCNASKFH QGALLVAVLP EYVIGTVAGG TGTEDTHPPY KQTQPGADGF ELQHPYVLDA G IPISQLTV CPHQWINLRT NNCATIIVPY INALPFDSAL NHCNFGLLVV PISPLDYDQG ATPVIPITIT LAPMCSEFAG LR QAVTQ UniProtKB: Genome polyprotein |
-Macromolecule #3: VP3
| Macromolecule | Name: VP3 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Enterovirus A71 Enterovirus A71 |
| Molecular weight | Theoretical: 26.468225 KDa |
| Recombinant expression | Organism:  Chlorocebus aethiops (grivet monkey) Chlorocebus aethiops (grivet monkey) |
| Sequence | String: GFPTELKPGT NQFLTTDDGV SAPILPNFHP TPCIHIPGEV RNLLELCQVE TILEVNNVPT NATSLMERLR FPVSAQAGKG ELCAVFRAD PGRNGPWQST LLGQLCGYYT QWSGSLEVTF MFTGSFMATG KMLIAYTPPG GPLPKDRATA MLGTHVIWDF G LQSSVTLV ...String: GFPTELKPGT NQFLTTDDGV SAPILPNFHP TPCIHIPGEV RNLLELCQVE TILEVNNVPT NATSLMERLR FPVSAQAGKG ELCAVFRAD PGRNGPWQST LLGQLCGYYT QWSGSLEVTF MFTGSFMATG KMLIAYTPPG GPLPKDRATA MLGTHVIWDF G LQSSVTLV IPWISNTHYR AHARDGVFDY YTTGLVSIWY QTNYVVPIGA PNTAYIIALA AAQKNFTMKL CKDASDILQT GT IQ UniProtKB: Genome polyprotein |
-Macromolecule #4: R10 ANTIBODY LIGHT CHAIN
| Macromolecule | Name: R10 ANTIBODY LIGHT CHAIN / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.283535 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DIVLTQSPAI MSASPGEKVT MTCSASSSVS YIHWYQQKSG TSPKRWIYDT SRLAFGVPGR FSGSGSGTSY SLTISSMEAE DAATYYCQQ WSSNYTFGGG TNLEIKRADA APTVSIFPPS SEQLTSGGAS VVCFLNNFYP KDINVKWKID GSERQNGVLN S WTDQDSKD ...String: DIVLTQSPAI MSASPGEKVT MTCSASSSVS YIHWYQQKSG TSPKRWIYDT SRLAFGVPGR FSGSGSGTSY SLTISSMEAE DAATYYCQQ WSSNYTFGGG TNLEIKRADA APTVSIFPPS SEQLTSGGAS VVCFLNNFYP KDINVKWKID GSERQNGVLN S WTDQDSKD STYSMSSTLT LTKDEYERHN SYTCEATHKT STSPIVKSFN RNEC |
-Macromolecule #5: R10 ANTIBODY HEAVY CHAIN
| Macromolecule | Name: R10 ANTIBODY HEAVY CHAIN / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 23.67165 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVKLVESGGG SVKPGGSLKL SCAASGFSFS TYGMSWVRQT PEKRLEWVAT ISGGGGYTYY PDSVKGRFTI SRDNARNILY LQMSSLRSG DTAMYYCARR VTTVAEYYFD YWGQGTTLTV SSPKTTPPSV YPLAPAAAQT NSMVTLGCLV KGYFPEPVTV T WNSGSLSS ...String: EVKLVESGGG SVKPGGSLKL SCAASGFSFS TYGMSWVRQT PEKRLEWVAT ISGGGGYTYY PDSVKGRFTI SRDNARNILY LQMSSLRSG DTAMYYCARR VTTVAEYYFD YWGQGTTLTV SSPKTTPPSV YPLAPAAAQT NSMVTLGCLV KGYFPEPVTV T WNSGSLSS GVHTFPAVLQ SDLYTLSSSV TVPSSTWPSE TVTCNVAHPA SSTKVDKKIV PR |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 20.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: INSILICO MODEL |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.9 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 1500 |
| Initial angle assignment | Type: RANDOM ASSIGNMENT |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)