+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6856 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human TRPC6 at 3.8A resolution | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRPC6 channel / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of ion transmembrane transporter activity / slit diaphragm / Role of second messengers in netrin-1 signaling / negative regulation of dendrite morphogenesis / store-operated calcium channel activity / Effects of PIP2 hydrolysis / Elevation of cytosolic Ca2+ levels / inositol 1,4,5 trisphosphate binding / positive regulation of calcium ion transport / cation channel complex ...positive regulation of ion transmembrane transporter activity / slit diaphragm / Role of second messengers in netrin-1 signaling / negative regulation of dendrite morphogenesis / store-operated calcium channel activity / Effects of PIP2 hydrolysis / Elevation of cytosolic Ca2+ levels / inositol 1,4,5 trisphosphate binding / positive regulation of calcium ion transport / cation channel complex / actinin binding / clathrin binding / TRP channels / monoatomic cation transport / regulation of cytosolic calcium ion concentration / single fertilization / monoatomic cation channel activity / positive regulation of neuron differentiation / calcium ion transmembrane transport / calcium channel activity / cellular response to hydrogen peroxide / neuron differentiation / actin binding / ATPase binding / positive regulation of cytosolic calcium ion concentration / cellular response to hypoxia / protein homodimerization activity / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Chen L / Tang Q | |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2018 Journal: Cell Res / Year: 2018Title: Structure of the receptor-activated human TRPC6 and TRPC3 ion channels. Authors: Qinglin Tang / Wenjun Guo / Li Zheng / Jing-Xiang Wu / Meng Liu / Xindi Zhou / Xiaolin Zhang / Lei Chen /  Abstract: TRPC6 and TRPC3 are receptor-activated nonselective cation channels that belong to the family of canonical transient receptor potential (TRPC) channels. They are activated by diacylglycerol, a lipid ...TRPC6 and TRPC3 are receptor-activated nonselective cation channels that belong to the family of canonical transient receptor potential (TRPC) channels. They are activated by diacylglycerol, a lipid second messenger. TRPC6 and TRPC3 are involved in many physiological processes and implicated in human genetic diseases. Here we present the structure of human TRPC6 homotetramer in complex with a newly identified high-affinity inhibitor BTDM solved by single-particle cryo-electron microscopy to 3.8 Å resolution. We also present the structure of human TRPC3 at 4.4 Å resolution. These structures show two-layer architectures in which the bell-shaped cytosolic layer holds the transmembrane layer. Extensive inter-subunit interactions of cytosolic domains, including the N-terminal ankyrin repeats and the C-terminal coiled-coil, contribute to the tetramer assembly. The high-affinity inhibitor BTDM wedges between the S5-S6 pore domain and voltage sensor-like domain to inhibit channel opening. Our structures uncover the molecular architecture of TRPC channels and provide a structural basis for understanding the mechanism of these channels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6856.map.gz emd_6856.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6856-v30.xml emd-6856-v30.xml emd-6856.xml emd-6856.xml | 9.5 KB 9.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6856.png emd_6856.png | 70.3 KB | ||
| Filedesc metadata |  emd-6856.cif.gz emd-6856.cif.gz | 5.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6856 http://ftp.pdbj.org/pub/emdb/structures/EMD-6856 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6856 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6856 | HTTPS FTP |
-Related structure data
| Related structure data |  5yx9MC  6911C  5zbgC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6856.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6856.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.33 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
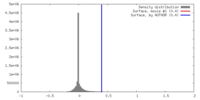
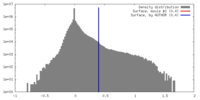
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : hTRPC6 homo-tetramer in complex with inhibitor
| Entire | Name: hTRPC6 homo-tetramer in complex with inhibitor |
|---|---|
| Components |
|
-Supramolecule #1: hTRPC6 homo-tetramer in complex with inhibitor
| Supramolecule | Name: hTRPC6 homo-tetramer in complex with inhibitor / type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Short transient receptor potential channel 6
| Macromolecule | Name: Short transient receptor potential channel 6 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 106.453242 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSQSPAFGPR RGSSPRGAAG AAARRNESQD YLLMDSELGE DGCPQAPLPC YGYYPCFRGS DNRLAHRRQT VLREKGRRLA NRGPAYMFS DRSTSLSIEE ERFLDAAEYG NIPVVRKMLE ECHSLNVNCV DYMGQNALQL AVANEHLEIT ELLLKKENLS R VGDALLLA ...String: MSQSPAFGPR RGSSPRGAAG AAARRNESQD YLLMDSELGE DGCPQAPLPC YGYYPCFRGS DNRLAHRRQT VLREKGRRLA NRGPAYMFS DRSTSLSIEE ERFLDAAEYG NIPVVRKMLE ECHSLNVNCV DYMGQNALQL AVANEHLEIT ELLLKKENLS R VGDALLLA ISKGYVRIVE AILSHPAFAE GKRLATSPSQ SELQQDDFYA YDEDGTRFSH DVTPIILAAH CQEYEIVHTL LR KGARIER PHDYFCKCND CNQKQKHDSF SHSRSRINAY KGLASPAYLS LSSEDPVMTA LELSNELAVL ANIEKEFKND YKK LSMQCK DFVVGLLDLC RNTEEVEAIL NGDVETLQSG DHGRPNLSRL KLAIKYEVKK FVAHPNCQQQ LLSIWYENLS GLRQ QTMAV KFLVVLAVAI GLPFLALIYW FAPCSKMGKI MRGPFMKFVA HAASFTIFLG LLVMNAADRF EGTKLLPNET STDNA KQLF RMKTSCFSWM EMLIISWVIG MIWAECKEIW TQGPKEYLFE LWNMLDFGML AIFAASFIAR FMAFWHASKA QSIIDA NDT LKDLTKVTLG DNVKYYNLAR IKWDPSDPQI ISEGLYAIAV VLSFSRIAYI LPANESFGPL QISLGRTVKD IFKFMVI FI MVFVAFMIGM FNLYSYYIGA KQNEAFTTVE ESFKTLFWAI FGLSEVKSVV INYNHKFIEN IGYVLYGVYN VTMVIVLL N MLIAMINSSF QEIEDDADVE WKFARAKLWF SYFEEGRTLP VPFNLVPSPK SLFYLLLKLK KWISELFQGH KKGFQEDAE MNKINEEKKL GILGSHEDLS KLSLDKKQVG HNKQPSIRSS EDFHLNSFNN PPRQYQKIMK RLIKRYVLQA QIDKESDEVN EGELKEIKQ DISSLRYELL EEKSQNTEDL AELIRELGEK LSMEPNQEET NR UniProtKB: Short transient receptor potential channel 6 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Applied symmetry - Point group: C4 (4 fold cyclic) / Resolution.type: BY AUTHOR / Resolution: 3.8 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC / Number images used: 53528 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)