+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6618 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the full-length TRPV2 channel by cryoEM | |||||||||
 Map data Map data | Structure of full-Length TRPV2 after signal subtraction 3D classification | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | TRPV2 / ion channel | |||||||||
| Function / homology |  Function and homology information Function and homology informationtransport / growth cone membrane / TRP channels / response to temperature stimulus / positive regulation of calcium ion import / calcium ion import across plasma membrane / positive regulation of axon extension / axonal growth cone / monoatomic cation channel activity / endomembrane system ...transport / growth cone membrane / TRP channels / response to temperature stimulus / positive regulation of calcium ion import / calcium ion import across plasma membrane / positive regulation of axon extension / axonal growth cone / monoatomic cation channel activity / endomembrane system / calcium channel activity / melanosome / lamellipodium / positive regulation of cold-induced thermogenesis / cell body / negative regulation of cell population proliferation / axon / neuronal cell body / cell surface / identical protein binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
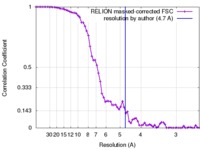
| Method | single particle reconstruction / cryo EM / Resolution: 4.7 Å | |||||||||
 Authors Authors | Huynh KW / Cohen MR / Jiang J / Samanta A / Lodowski DT / Zhou ZH / Moiseenkova-Bell VY | |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2016 Journal: Nat Commun / Year: 2016Title: Structure of the full-length TRPV2 channel by cryo-EM. Authors: Kevin W Huynh / Matthew R Cohen / Jiansen Jiang / Amrita Samanta / David T Lodowski / Z Hong Zhou / Vera Y Moiseenkova-Bell /  Abstract: Transient receptor potential (TRP) proteins form a superfamily Ca(2+)-permeable cation channels regulated by a range of chemical and physical stimuli. Structural analysis of a 'minimal' TRP vanilloid ...Transient receptor potential (TRP) proteins form a superfamily Ca(2+)-permeable cation channels regulated by a range of chemical and physical stimuli. Structural analysis of a 'minimal' TRP vanilloid subtype 1 (TRPV1) elucidated a mechanism of channel activation by agonists through changes in its outer pore region. Though homologous to TRPV1, other TRPV channels (TRPV2-6) are insensitive to TRPV1 activators including heat and vanilloids. To further understand the structural basis of TRPV channel function, we determined the structure of full-length TRPV2 at ∼5 Å resolution by cryo-electron microscopy. Like TRPV1, TRPV2 contains two constrictions, one each in the pore-forming upper and lower gates. The agonist-free full-length TRPV2 has wider upper and lower gates compared with closed and agonist-activated TRPV1. We propose these newly revealed TRPV2 structural features contribute to diversity of TRPV channels. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6618.map.gz emd_6618.map.gz | 25.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6618-v30.xml emd-6618-v30.xml emd-6618.xml emd-6618.xml | 10.5 KB 10.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6618_fsc.xml emd_6618_fsc.xml | 6.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_6618.png emd_6618.png | 86.4 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6618 http://ftp.pdbj.org/pub/emdb/structures/EMD-6618 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6618 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6618 | HTTPS FTP |
-Related structure data
| Related structure data |  5hi9MC  6580C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6618.map.gz / Format: CCP4 / Size: 26.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6618.map.gz / Format: CCP4 / Size: 26.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of full-Length TRPV2 after signal subtraction 3D classification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.29 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Recombinant rat full-length TRPV2
| Entire | Name: Recombinant rat full-length TRPV2 |
|---|---|
| Components |
|
-Supramolecule #1000: Recombinant rat full-length TRPV2
| Supramolecule | Name: Recombinant rat full-length TRPV2 / type: sample / ID: 1000 / Details: The sample was monodisperse / Oligomeric state: tetramer / Number unique components: 1 |
|---|---|
| Molecular weight | Experimental: 600 KDa / Theoretical: 340 KDa / Method: gel filtration |
-Macromolecule #1: Transient Receptor Potential Cation Channel, Subfamily V, Member 2
| Macromolecule | Name: Transient Receptor Potential Cation Channel, Subfamily V, Member 2 type: protein_or_peptide / ID: 1 / Name.synonym: TRPV2 / Number of copies: 4 / Oligomeric state: tetramer / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Experimental: 600 KDa / Theoretical: 340 KDa |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: Transient receptor potential cation channel subfamily V member 2 GO: transport InterPro: Transient receptor potential cation channel subfamily V member 2 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 8 Details: 0.064 mM DMNG, 150 mM NaCl, 20 mM HEPES, 1.0 mM DTT |
| Grid | Details: Quantifoil R2/1 400 mesh copper grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 120 K / Instrument: HOMEMADE PLUNGER / Method: Blotted twice. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 100,000 times magnification. |
| Date | Jan 12, 2014 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number real images: 988 Details: Every image is the average of 14 frames recorded by the direct electron detector. |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 31000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)