[English] 日本語
 Yorodumi
Yorodumi- EMDB-6535: Structure of the eukaryotic replicative CMG helicase and pumpjack... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6535 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of the eukaryotic replicative CMG helicase and pumpjack motion | |||||||||
 Map data Map data | Structure of CMG with "Flat" Mcm2-7 ring | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CMG helicase / Cryo-EM | |||||||||
| Function / homology |  Function and homology information Function and homology informationUnwinding of DNA / MCM core complex / Assembly of the pre-replicative complex / Switching of origins to a post-replicative state / DNA strand elongation involved in mitotic DNA replication / GINS complex / MCM complex binding / mitotic DNA replication preinitiation complex assembly / nuclear DNA replication / premeiotic DNA replication ...Unwinding of DNA / MCM core complex / Assembly of the pre-replicative complex / Switching of origins to a post-replicative state / DNA strand elongation involved in mitotic DNA replication / GINS complex / MCM complex binding / mitotic DNA replication preinitiation complex assembly / nuclear DNA replication / premeiotic DNA replication / replication fork protection complex / pre-replicative complex assembly involved in nuclear cell cycle DNA replication / Activation of the pre-replicative complex / mitotic DNA replication / CMG complex / nuclear pre-replicative complex / DNA replication preinitiation complex / Activation of ATR in response to replication stress / MCM complex / double-strand break repair via break-induced replication / mitotic DNA replication initiation / single-stranded DNA helicase activity / silent mating-type cassette heterochromatin formation / regulation of DNA-templated DNA replication initiation / DNA strand elongation involved in DNA replication / nuclear replication fork / DNA replication origin binding / DNA replication initiation / subtelomeric heterochromatin formation / DNA helicase activity / transcription elongation by RNA polymerase II / helicase activity / DNA-templated DNA replication / heterochromatin formation / single-stranded DNA binding / DNA helicase / DNA replication / chromosome, telomeric region / DNA damage response / chromatin binding / ATP hydrolysis activity / zinc ion binding / nucleoplasm / ATP binding / nucleus / cytoplasm Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.7 Å | |||||||||
 Authors Authors | Yuan Z / Bai L / Sun J / Georgescu RE / Liu J / O'Donnell ME / Li H | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2016 Journal: Nat Struct Mol Biol / Year: 2016Title: Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Authors: Zuanning Yuan / Lin Bai / Jingchuan Sun / Roxana Georgescu / Jun Liu / Michael E O'Donnell / Huilin Li /  Abstract: The CMG helicase is composed of Cdc45, Mcm2-7 and GINS. Here we report the structure of the Saccharomyces cerevisiae CMG, determined by cryo-EM at a resolution of 3.7-4.8 Å. The structure reveals ...The CMG helicase is composed of Cdc45, Mcm2-7 and GINS. Here we report the structure of the Saccharomyces cerevisiae CMG, determined by cryo-EM at a resolution of 3.7-4.8 Å. The structure reveals that GINS and Cdc45 scaffold the N tier of the helicase while enabling motion of the AAA+ C tier. CMG exists in two alternating conformations, compact and extended, thus suggesting that the helicase moves like an inchworm. The N-terminal regions of Mcm2-7, braced by Cdc45-GINS, form a rigid platform upon which the AAA+ C domains make longitudinal motions, nodding up and down like an oil-rig pumpjack attached to a stable platform. The Mcm ring is remodeled in CMG relative to the inactive Mcm2-7 double hexamer. The Mcm5 winged-helix domain is inserted into the central channel, thus blocking entry of double-stranded DNA and supporting a steric-exclusion DNA-unwinding model. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6535.map.gz emd_6535.map.gz | 59.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6535-v30.xml emd-6535-v30.xml emd-6535.xml emd-6535.xml | 9.8 KB 9.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6535.jpg emd_6535.jpg | 169 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6535 http://ftp.pdbj.org/pub/emdb/structures/EMD-6535 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6535 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6535 | HTTPS FTP |
-Related structure data
| Related structure data |  3jc5MC  6534C  6536C  3jc6C  3jc7C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6535.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6535.map.gz / Format: CCP4 / Size: 62.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Structure of CMG with "Flat" Mcm2-7 ring | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.01 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
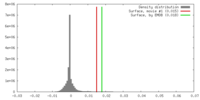
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Saccharomyces cerevisiae CMG complex
| Entire | Name: Saccharomyces cerevisiae CMG complex |
|---|---|
| Components |
|
-Supramolecule #1000: Saccharomyces cerevisiae CMG complex
| Supramolecule | Name: Saccharomyces cerevisiae CMG complex / type: sample / ID: 1000 / Number unique components: 1 |
|---|---|
| Molecular weight | Theoretical: 700 KDa |
-Macromolecule #1: CMG complex
| Macromolecule | Name: CMG complex / type: protein_or_peptide / ID: 1 / Name.synonym: CMG / Number of copies: 1 / Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 700 KDa |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | Details: 20 mM Tris acetate, 40 mM potassium glutamate, 2 mM DTT, 0.1 mM EDTA |
| Grid | Details: 400 mesh holey carbon C-flat grid, glow-discharged in air |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: FEI VITROBOT MARK IV / Method: Blot for 3 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Date | Aug 1, 2015 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Number real images: 8000 / Average electron dose: 50 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 49505 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Details | All steps, including automatic particle picking, 2D classification, 3D classification, and 3D refinement were performed in Relion 1.4. |
|---|---|
| CTF correction | Details: CTFFIND4 |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 4.7 Å / Resolution method: OTHER / Software - Name: Relion_1.4 / Number images used: 178530 |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)