[English] 日本語
 Yorodumi
Yorodumi- EMDB-6458: Cryo-EM Structure of the Activated NAIP2/NLRC4 Inflammasome Revea... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6458 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM Structure of the Activated NAIP2/NLRC4 Inflammasome Reveals Nucleated Polymerization | |||||||||
 Map data Map data | Reconstruction of 11-fold NAIP2/NLRC4 inflammasome | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Inflammasome / NLRC4 / NAIP2 | |||||||||
| Function / homology |  Function and homology information Function and homology informationIPAF inflammasome complex / caspase binding / positive regulation of protein processing / pyroptotic inflammatory response / detection of bacterium / endopeptidase activator activity / activation of innate immune response / positive regulation of interleukin-1 beta production / protein homooligomerization / regulation of apoptotic process ...IPAF inflammasome complex / caspase binding / positive regulation of protein processing / pyroptotic inflammatory response / detection of bacterium / endopeptidase activator activity / activation of innate immune response / positive regulation of interleukin-1 beta production / protein homooligomerization / regulation of apoptotic process / defense response to bacterium / positive regulation of apoptotic process / inflammatory response / innate immune response / intracellular membrane-bounded organelle / apoptotic process / protein homodimerization activity / ATP binding / identical protein binding / plasma membrane / cytosol Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.7 Å | |||||||||
 Authors Authors | Zhang L / Chen S / Ruan J / Wu J / Tong AB / Yin Q / Li Y / David L / Lu A / Wang WL ...Zhang L / Chen S / Ruan J / Wu J / Tong AB / Yin Q / Li Y / David L / Lu A / Wang WL / Marks C / Ouyang Q / Zhang X / Mao Y / Wu H | |||||||||
 Citation Citation |  Journal: Science / Year: 2015 Journal: Science / Year: 2015Title: Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Authors: Liman Zhang / Shuobing Chen / Jianbin Ruan / Jiayi Wu / Alexander B Tong / Qian Yin / Yang Li / Liron David / Alvin Lu / Wei Li Wang / Carolyn Marks / Qi Ouyang / Xinzheng Zhang / Youdong Mao / Hao Wu /   Abstract: The NLR family apoptosis inhibitory proteins (NAIPs) bind conserved bacterial ligands, such as the bacterial rod protein PrgJ, and recruit NLR family CARD-containing protein 4 (NLRC4) as the ...The NLR family apoptosis inhibitory proteins (NAIPs) bind conserved bacterial ligands, such as the bacterial rod protein PrgJ, and recruit NLR family CARD-containing protein 4 (NLRC4) as the inflammasome adapter to activate innate immunity. We found that the PrgJ-NAIP2-NLRC4 inflammasome is assembled into multisubunit disk-like structures through a unidirectional adenosine triphosphatase polymerization, primed with a single PrgJ-activated NAIP2 per disk. Cryo-electron microscopy (cryo-EM) reconstruction at subnanometer resolution revealed a ~90° hinge rotation accompanying NLRC4 activation. Unlike in the related heptameric Apaf-1 apoptosome, in which each subunit needs to be conformationally activated by its ligand before assembly, a single PrgJ-activated NAIP2 initiates NLRC4 polymerization in a domino-like reaction to promote the disk assembly. These insights reveal the mechanism of signal amplification in NAIP-NLRC4 inflammasomes. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6458.map.gz emd_6458.map.gz | 313.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6458-v30.xml emd-6458-v30.xml emd-6458.xml emd-6458.xml | 13.2 KB 13.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_6458.png emd_6458.png | 322.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6458 http://ftp.pdbj.org/pub/emdb/structures/EMD-6458 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6458 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6458 | HTTPS FTP |
-Related structure data
| Related structure data |  3jblMC  6459C  6460C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | |
| EM raw data |  EMPIAR-10063 (Title: Cryo-EM Structure of the Activated NAIP2/NLRC4 Inflammasome Reveals Nucleated Polymerization EMPIAR-10063 (Title: Cryo-EM Structure of the Activated NAIP2/NLRC4 Inflammasome Reveals Nucleated PolymerizationData size: 1.7 TB Data #1: Drift-corrected micrographs of NLRC4/NAIP2 inflammasome [micrographs - single frame] Data #2: Single-particle stacks of NLRC4/NAIP2 inflammasome [picked particles - multiframe - processed]) |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6458.map.gz / Format: CCP4 / Size: 335 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6458.map.gz / Format: CCP4 / Size: 335 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of 11-fold NAIP2/NLRC4 inflammasome | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.86 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
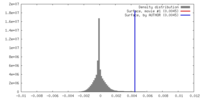
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : NAIP2/NLRC4 inflammasome, 11-fold disk
| Entire | Name: NAIP2/NLRC4 inflammasome, 11-fold disk |
|---|---|
| Components |
|
-Supramolecule #1000: NAIP2/NLRC4 inflammasome, 11-fold disk
| Supramolecule | Name: NAIP2/NLRC4 inflammasome, 11-fold disk / type: sample / ID: 1000 Details: The sample contains multiple oligomeric states, including 10-fold, 11-fold, and 12-fold disks. Oligomeric state: One NAIP2 subunit and ten NLRC subunits / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 1.8 MDa / Theoretical: 1.8 MDa |
-Macromolecule #1: NLR family CARD domain-containing protein 4
| Macromolecule | Name: NLR family CARD domain-containing protein 4 / type: protein_or_peptide / ID: 1 / Name.synonym: NLRC4 Details: One activated NAIP2 induces polymerization of NLRC4. Number of copies: 10 Oligomeric state: NLRC4 inflammasome comprises ten NLRC4 and one NAIP2 Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Tissue: Abelson murine leukemia virus-induced tumor; ascites Cell: macrophage / Location in cell: cytosol |
| Recombinant expression | Organism:  |
| Sequence | UniProtKB: NLR family CARD domain-containing protein 4 |
-Macromolecule #2: NLR family, apoptosis inhibitory protein 2
| Macromolecule | Name: NLR family, apoptosis inhibitory protein 2 / type: protein_or_peptide / ID: 2 / Name.synonym: NAIP2 Details: One activated NAIP2 induces polymerization of NLRC4. Number of copies: 1 Oligomeric state: NLRC4 inflammasome comprises one NAIP2 and ten NLRC4 Recombinant expression: Yes |
|---|---|
| Source (natural) | Organism:  Tissue: Abelson murine leukemia virus-induced tumor; ascites Cell: macrophage / Location in cell: cytosol |
| Recombinant expression | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 2 mM DTT |
| Grid | Details: 400 mesh C-flat grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 103 K / Instrument: FEI VITROBOT MARK IV / Method: Blot for one second before plunging |
- Electron microscopy
Electron microscopy
| Microscope | OTHER |
|---|---|
| Temperature | Min: 79.5 K / Max: 80 K / Average: 79.5 K |
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 21,000 times magnification. |
| Date | Apr 1, 2015 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Digitization - Sampling interval: 5 µm / Number real images: 9113 / Average electron dose: 48 e/Å2 Details: average of 36 frames recorded by the direct electron detector Bits/pixel: 8 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 28736 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 21000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
- Image processing
Image processing
| CTF correction | Details: Wiener-type filter |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 4.7 Å / Resolution method: OTHER / Software - Name: Spider, EMAN2, Relion / Number images used: 75114 |
 Movie
Movie Controller
Controller













 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)