[English] 日本語
 Yorodumi
Yorodumi- EMDB-6375: A putative ATPase mediates RNA transcription and capping in a dsR... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6375 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | A putative ATPase mediates RNA transcription and capping in a dsRNA virus | |||||||||
 Map data Map data | Reconstruction of S-CPV | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Viral ATPase / histidine-mediated guanylyl transfer / conformational changes / regulation of transcription | |||||||||
| Function / homology |  Function and homology information Function and homology information | |||||||||
| Biological species |   Bombyx mori cypovirus 1 Bombyx mori cypovirus 1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Yu XK / Jiang JS / Sun JC / Zhou ZH | |||||||||
 Citation Citation |  Journal: Elife / Year: 2015 Journal: Elife / Year: 2015Title: A putative ATPase mediates RNA transcription and capping in a dsRNA virus. Authors: Xuekui Yu / Jiansen Jiang / Jingchen Sun / Z Hong Zhou /  Abstract: mRNA transcription in dsRNA viruses is a highly regulated process but the mechanism of this regulation is not known. Here, by nucleoside triphosphatase (NTPase) assay and comparisons of six high- ...mRNA transcription in dsRNA viruses is a highly regulated process but the mechanism of this regulation is not known. Here, by nucleoside triphosphatase (NTPase) assay and comparisons of six high-resolution (2.9-3.1 Å) cryo-electron microscopy structures of cytoplasmic polyhedrosis virus with bound ligands, we show that the large sub-domain of the guanylyltransferase (GTase) domain of the turret protein (TP) also has an ATP-binding site and is likely an ATPase. S-adenosyl-L-methionine (SAM) acts as a signal and binds the methylase-2 domain of TP to induce conformational change of the viral capsid, which in turn activates the putative ATPase. ATP binding/hydrolysis leads to an enlarged capsid for efficient mRNA synthesis, an open GTase domain for His217-mediated guanylyl transfer, and an open methylase-1 domain for SAM binding and methyl transfer. Taken together, our data support a role of the putative ATPase in mediating the activation of mRNA transcription and capping within the confines of the virus. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6375.map.gz emd_6375.map.gz | 568.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6375-v30.xml emd-6375-v30.xml emd-6375.xml emd-6375.xml | 9.6 KB 9.6 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6375.gif 400_6375.gif 80_6375.gif 80_6375.gif | 77.2 KB 3.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6375 http://ftp.pdbj.org/pub/emdb/structures/EMD-6375 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6375 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6375 | HTTPS FTP |
-Related structure data
| Related structure data |  3jb1MC  6371C  6374C  6376C  6377C  6378C  3jayC  3jazC  3jb0C  3jb2C  3jb3C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6375.map.gz / Format: CCP4 / Size: 1.2 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6375.map.gz / Format: CCP4 / Size: 1.2 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of S-CPV | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.104 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
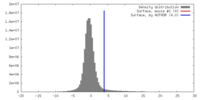
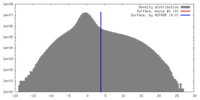
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Cytoplasmic polyhedrosis virus with SAM
| Entire | Name: Cytoplasmic polyhedrosis virus with SAM |
|---|---|
| Components |
|
-Supramolecule #1000: Cytoplasmic polyhedrosis virus with SAM
| Supramolecule | Name: Cytoplasmic polyhedrosis virus with SAM / type: sample / ID: 1000 / Oligomeric state: icosahedral / Number unique components: 1 |
|---|
-Supramolecule #1: Bombyx mori cypovirus 1
| Supramolecule | Name: Bombyx mori cypovirus 1 / type: virus / ID: 1 / NCBI-ID: 110829 / Sci species name: Bombyx mori cypovirus 1 / Database: NCBI / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
| Virus shell | Shell ID: 1 / Diameter: 720 Å / T number (triangulation number): 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK II |
|---|
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 135,000 times magnification. |
| Date | May 8, 2012 |
| Image recording | Category: FILM / Film or detector model: KODAK SO-163 FILM / Digitization - Scanner: NIKON SUPER COOLSCAN 9000 / Average electron dose: 25 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 60535 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.75 mm / Nominal magnification: 59000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: Each particle |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: OTHER / Software - Name: IMIRS / Number images used: 44908 |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)