[English] 日本語
 Yorodumi
Yorodumi- EMDB-6240: Atomic structure of a non-enveloped virus reveals pH sensors for ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6240 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Atomic structure of a non-enveloped virus reveals pH sensors for a coordinated process of cell entry | |||||||||
 Map data Map data | The averaged density from the reconstruction of BTV virion | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Non-enveloped virus / cell entry / cryo-EM / pH sensor | |||||||||
| Function / homology | Outer capsid protein VP5, Orbivirus / Orbivirus outer capsid protein VP5 / viral outer capsid / symbiont entry into host cell via permeabilization of host membrane / structural molecule activity / Outer capsid protein VP5 Function and homology information Function and homology information | |||||||||
| Biological species |  Bluetongue virus 1 Bluetongue virus 1 | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Zhang X / Patel A / Celma C / Roy P / Zhou ZH | |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2016 Journal: Nat Struct Mol Biol / Year: 2016Title: Atomic model of a nonenveloped virus reveals pH sensors for a coordinated process of cell entry. Authors: Xing Zhang / Avnish Patel / Cristina C Celma / Xuekui Yu / Polly Roy / Z Hong Zhou /   Abstract: Viruses sense environmental cues such as pH to engage in membrane interactions for cell entry during infection, but how nonenveloped viruses sense pH is largely undefined. Here, we report both high- ...Viruses sense environmental cues such as pH to engage in membrane interactions for cell entry during infection, but how nonenveloped viruses sense pH is largely undefined. Here, we report both high- and low-pH structures of bluetongue virus (BTV), which enters cells via a two-stage endosomal process. The receptor-binding protein VP2 possesses a zinc finger that may function to maintain VP2 in a metastable state and a conserved His866, which senses early-endosomal pH. The membrane-penetration protein VP5 has three domains: dagger, unfurling and anchoring. Notably, the β-meander motif of the anchoring domain contains a histidine cluster that can sense late-endosomal pH and also possesses four putative membrane-interaction elements. Exposing BTV to low pH detaches VP2 and dramatically refolds the dagger and unfurling domains of VP5. Our biochemical and structure-guided-mutagenesis studies support these coordinated pH-sensing mechanisms. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6240.map.gz emd_6240.map.gz | 283.6 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6240-v30.xml emd-6240-v30.xml emd-6240.xml emd-6240.xml | 9.2 KB 9.2 KB | Display Display |  EMDB header EMDB header |
| Images |  400_6240.gif 400_6240.gif 80_6240.gif 80_6240.gif | 42.1 KB 3.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6240 http://ftp.pdbj.org/pub/emdb/structures/EMD-6240 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6240 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6240 | HTTPS FTP |
-Related structure data
| Related structure data |  3j9eMC  6239C  6444C  6445C  3j9dC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6240.map.gz / Format: CCP4 / Size: 8.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6240.map.gz / Format: CCP4 / Size: 8.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | The averaged density from the reconstruction of BTV virion | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.01 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
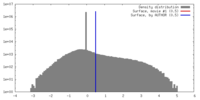
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : VP5 of Bluetongue Virus
| Entire | Name: VP5 of Bluetongue Virus |
|---|---|
| Components |
|
-Supramolecule #1000: VP5 of Bluetongue Virus
| Supramolecule | Name: VP5 of Bluetongue Virus / type: sample / ID: 1000 / Number unique components: 1 |
|---|
-Supramolecule #1: Bluetongue virus 1
| Supramolecule | Name: Bluetongue virus 1 / type: virus / ID: 1 / NCBI-ID: 35327 / Sci species name: Bluetongue virus 1 / Database: NCBI / Virus type: VIRION / Virus isolate: SPECIES / Virus enveloped: No / Virus empty: No |
|---|---|
| Host (natural) | Organism:  |
| Virus shell | Shell ID: 1 / Diameter: 800 Å / T number (triangulation number): 13 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8.8 |
|---|---|
| Grid | Details: 400 mesh grid with thin carbon support |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 90 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Average: 82 K |
| Date | Nov 29, 2013 |
| Image recording | Category: CCD / Film or detector model: GATAN K2 (4k x 4k) / Digitization - Sampling interval: 5 µm / Number real images: 1630 / Average electron dose: 20 e/Å2 / Bits/pixel: 32 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 24140 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.3 µm / Nominal defocus min: 0.7 µm / Nominal magnification: 14000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Details: each particle |
|---|---|
| Final reconstruction | Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 3.3 Å / Resolution method: OTHER / Software - Name: Frealign / Number images used: 5008 |
| Final two d classification | Number classes: 1 |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)