[English] 日本語
 Yorodumi
Yorodumi- PDB-4v68: T. thermophilus 70S ribosome in complex with mRNA, tRNAs and EF-T... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4v68 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | T. thermophilus 70S ribosome in complex with mRNA, tRNAs and EF-Tu.GDP.kirromycin ternary complex, fitted to a 6.4 A Cryo-EM map. | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords | RIBOSOME / cryo-electron microscopy/elongation factor/GTPase/ribosome/translation / Ribonucleoprotein / Ribosomal protein / RNA-binding / rRNA-binding / Metal-binding / Zinc-finger / tRNA-binding / Elongation factor / GTP-binding / Nucleotide-binding / Phosphoprotein / Protein biosynthesis | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein-synthesizing GTPase / translation elongation factor activity / regulation of translation / large ribosomal subunit / transferase activity / ribosomal small subunit biogenesis / ribosomal small subunit assembly / ribosomal large subunit assembly / 5S rRNA binding / small ribosomal subunit ...protein-synthesizing GTPase / translation elongation factor activity / regulation of translation / large ribosomal subunit / transferase activity / ribosomal small subunit biogenesis / ribosomal small subunit assembly / ribosomal large subunit assembly / 5S rRNA binding / small ribosomal subunit / small ribosomal subunit rRNA binding / cytosolic small ribosomal subunit / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding / structural constituent of ribosome / ribosome / translation / ribonucleoprotein complex / GTPase activity / mRNA binding / GTP binding / RNA binding / zinc ion binding / metal ion binding / cytoplasm / cytosol Similarity search - Function | |||||||||
| Biological species |   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) | |||||||||
| Method | ELECTRON MICROSCOPY / single particle reconstruction / cryo EM / Resolution: 6.4 Å | |||||||||
 Authors Authors | Schuette, J.-C. / Spahn, C.M.T. | |||||||||
 Citation Citation |  Journal: EMBO J / Year: 2009 Journal: EMBO J / Year: 2009Title: GTPase activation of elongation factor EF-Tu by the ribosome during decoding. Authors: Jan-Christian Schuette / Frank V Murphy / Ann C Kelley / John R Weir / Jan Giesebrecht / Sean R Connell / Justus Loerke / Thorsten Mielke / Wei Zhang / Pawel A Penczek / V Ramakrishnan / Christian M T Spahn /  Abstract: We have used single-particle reconstruction in cryo-electron microscopy to determine a structure of the Thermus thermophilus ribosome in which the ternary complex of elongation factor Tu (EF-Tu), ...We have used single-particle reconstruction in cryo-electron microscopy to determine a structure of the Thermus thermophilus ribosome in which the ternary complex of elongation factor Tu (EF-Tu), tRNA and guanine nucleotide has been trapped on the ribosome using the antibiotic kirromycin. This represents the state in the decoding process just after codon recognition by tRNA and the resulting GTP hydrolysis by EF-Tu, but before the release of EF-Tu from the ribosome. Progress in sample purification and image processing made it possible to reach a resolution of 6.4 A. Secondary structure elements in tRNA, EF-Tu and the ribosome, and even GDP and kirromycin, could all be visualized directly. The structure reveals a complex conformational rearrangement of the tRNA in the A/T state and the interactions with the functionally important switch regions of EF-Tu crucial to GTP hydrolysis. Thus, the structure provides insights into the molecular mechanism of signalling codon recognition from the decoding centre of the 30S subunit to the GTPase centre of EF-Tu. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4v68.cif.gz 4v68.cif.gz | 3.6 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4v68.ent.gz pdb4v68.ent.gz | Display |  PDB format PDB format | |
| PDBx/mmJSON format |  4v68.json.gz 4v68.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/v6/4v68 https://data.pdbj.org/pub/pdb/validation_reports/v6/4v68 ftp://data.pdbj.org/pub/pdb/validation_reports/v6/4v68 ftp://data.pdbj.org/pub/pdb/validation_reports/v6/4v68 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5030MC M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
|
|---|---|
| 1 |
|
- Components
Components
-RNA chain , 8 types, 8 molecules AAAVAXAWAYA0BABB
| #1: RNA chain | Mass: 488391.188 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
|---|---|
| #22: RNA chain | Mass: 24816.811 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| #23: RNA chain | Mass: 1923.237 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: Chemically synthesized |
| #24: RNA chain | Mass: 24485.539 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| #25: RNA chain | Mass: 23583.344 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| #26: RNA chain | Mass: 873.540 Da / Num. of mol.: 1 / Source method: obtained synthetically / Details: Chemically synthesized / References: UniProt: Q5SHN6 |
| #38: RNA chain | Mass: 927659.562 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
| #39: RNA chain | Mass: 38553.000 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) Thermus thermophilus (bacteria) |
-30S ribosomal protein ... , 20 types, 20 molecules ABADAEAFAHAKALAOAPAQARATACAGAIAJAMANASAU
| #2: Protein | Mass: 27116.385 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P80371 Thermus thermophilus (bacteria) / References: UniProt: P80371 |
|---|---|
| #3: Protein | Mass: 24242.254 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P80373 Thermus thermophilus (bacteria) / References: UniProt: P80373 |
| #4: Protein | Mass: 16460.193 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: Q5SHQ5 Thermus thermophilus (bacteria) / References: UniProt: Q5SHQ5 |
| #5: Protein | Mass: 11988.753 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: Q5SLP8 Thermus thermophilus (bacteria) / References: UniProt: Q5SLP8 |
| #6: Protein | Mass: 15868.570 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: Q5SHQ2, UniProt: A0A0M9AFS9*PLUS Thermus thermophilus (bacteria) / References: UniProt: Q5SHQ2, UniProt: A0A0M9AFS9*PLUS |
| #7: Protein | Mass: 12606.369 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P80376 Thermus thermophilus (bacteria) / References: UniProt: P80376 |
| #8: Protein | Mass: 13875.388 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: Q5SHN3 Thermus thermophilus (bacteria) / References: UniProt: Q5SHN3 |
| #9: Protein | Mass: 10447.213 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: Q5SJ76 Thermus thermophilus (bacteria) / References: UniProt: Q5SJ76 |
| #10: Protein | Mass: 9995.546 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: Q5SJH3 Thermus thermophilus (bacteria) / References: UniProt: Q5SJH3 |
| #11: Protein | Mass: 11880.098 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: Q5SHP7, UniProt: A0A0N0BLS5*PLUS Thermus thermophilus (bacteria) / References: UniProt: Q5SHP7, UniProt: A0A0N0BLS5*PLUS |
| #12: Protein | Mass: 8155.812 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: Q5SLQ0 Thermus thermophilus (bacteria) / References: UniProt: Q5SLQ0 |
| #13: Protein | Mass: 10921.086 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P80380 Thermus thermophilus (bacteria) / References: UniProt: P80380 |
| #14: Protein | Mass: 22975.588 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P80372 Thermus thermophilus (bacteria) / References: UniProt: P80372 |
| #15: Protein | Mass: 17919.775 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P17291 Thermus thermophilus (bacteria) / References: UniProt: P17291 |
| #16: Protein | Mass: 14298.466 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P62669 Thermus thermophilus (bacteria) / References: UniProt: P62669 |
| #17: Protein | Mass: 11398.308 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: Q5SHN7 Thermus thermophilus (bacteria) / References: UniProt: Q5SHN7 |
| #18: Protein | Mass: 14207.666 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P80377 Thermus thermophilus (bacteria) / References: UniProt: P80377 |
| #19: Protein | Mass: 7027.529 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: Q5SHQ1, UniProt: A0A0N0BLP2*PLUS Thermus thermophilus (bacteria) / References: UniProt: Q5SHQ1, UniProt: A0A0N0BLP2*PLUS |
| #20: Protein | Mass: 9006.486 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: Q5SHP2 Thermus thermophilus (bacteria) / References: UniProt: Q5SHP2 |
| #21: Protein/peptide | Mass: 3089.655 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: Q5SIH3 Thermus thermophilus (bacteria) / References: UniProt: Q5SIH3 |
-Protein , 1 types, 1 molecules AZ
| #27: Protein | Mass: 44709.887 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Thermus thermophilus (bacteria) / References: UniProt: P60339 Thermus thermophilus (bacteria) / References: UniProt: P60339 |
|---|
+50S ribosomal protein ... , 31 types, 31 molecules B0B1B2B3B4B5B6B7B8B9BCBDBEBFBGBHBIBLBNBOBPBQBRBSBTBUBVBWBXBYBZ
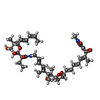
-Non-polymers , 4 types, 4 molecules 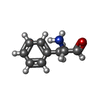





| #61: Chemical | ChemComp-PHA / |
|---|---|
| #62: Chemical | ChemComp-GDP / |
| #63: Chemical | ChemComp-MAU / |
| #64: Chemical | ChemComp-BME / |
-Details
| Has protein modification | Y |
|---|
-Experimental details
-Experiment
| Experiment | Method: ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method: single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Thermus thermophilus ribosome complexed with mRNA, tRNAs and EF-Tu Type: RIBOSOME / Details: 70S tRNA EF-Tu GDP kirromycin ribosomal complexes |
|---|---|
| Specimen | Conc.: 30 mg/ml / Embedding applied: NO / Shadowing applied: NO / Staining applied: NO / Vitrification applied: YES |
| Specimen support | Details: Quantifoil grids. |
| Vitrification | Instrument: FEI VITROBOT MARK I / Cryogen name: METHANE Details: A Vitrobot was used to shock-freeze the sample in liquid methane. |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI POLARA 300 |
| Electron gun | Electron source:  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: SPOT SCAN |
| Electron lens | Mode: BRIGHT FIELD / Nominal magnification: 39000 X / Nominal defocus max: 4500 nm / Nominal defocus min: 1500 nm |
| Specimen holder | Temperature: 77 K |
| Image recording | Electron dose: 23 e/Å2 / Film or detector model: KODAK SO-163 FILM |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Relative weight: 1 |
- Processing
Processing
| CTF correction | Details: CTF correction of each defocus group volume prior to back projection. | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Symmetry | Point symmetry: C1 (asymmetric) | ||||||||||||
| 3D reconstruction | Method: projection matching / Resolution: 6.4 Å / Num. of particles: 323688 / Nominal pixel size: 1.26 Å / Actual pixel size: 1.262 Å / Details: The SPIDER software package was used. / Symmetry type: POINT | ||||||||||||
| Atomic model building | Protocol: RIGID BODY FIT / Space: REAL Target criteria: best visual fit using the programs O, CHIMERA or PYMOL. Details: REFINEMENT PROTOCOL--rigid body docking carried out with SPIDER or SITUS software. | ||||||||||||
| Refinement step | Cycle: LAST
|
 Movie
Movie Controller
Controller












 PDBj
PDBj




































