[English] 日本語
 Yorodumi
Yorodumi- PDB-4cox: CYCLOOXYGENASE-2 (PROSTAGLANDIN SYNTHASE-2) COMPLEXED WITH A NON-... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4cox | ||||||
|---|---|---|---|---|---|---|---|
| Title | CYCLOOXYGENASE-2 (PROSTAGLANDIN SYNTHASE-2) COMPLEXED WITH A NON-SELECTIVE INHIBITOR, INDOMETHACIN | ||||||
 Components Components | CYCLOOXYGENASE-2 | ||||||
 Keywords Keywords | OXIDOREDUCTASE / PEROXIDASE / DIOXYGENASE / CYCLOOXYGENASE / NONSTEROIDAL ANTIINFLAMMATORY DRUGS / INFLAMMATION / ARTHRITIS / PROSTAGLANDIN / PROSTAGLANDIN SYNTHASE | ||||||
| Function / homology |  Function and homology information Function and homology informationBiosynthesis of DHA-derived SPMs / Biosynthesis of EPA-derived SPMs / Biosynthesis of DPAn-3 SPMs / Biosynthesis of electrophilic ω-3 PUFA oxo-derivatives / Synthesis of 15-eicosatetraenoic acid derivatives / cellular response to non-ionic osmotic stress / : / Synthesis of Prostaglandins (PG) and Thromboxanes (TX) / prostaglandin-endoperoxide synthase / prostaglandin-endoperoxide synthase activity ...Biosynthesis of DHA-derived SPMs / Biosynthesis of EPA-derived SPMs / Biosynthesis of DPAn-3 SPMs / Biosynthesis of electrophilic ω-3 PUFA oxo-derivatives / Synthesis of 15-eicosatetraenoic acid derivatives / cellular response to non-ionic osmotic stress / : / Synthesis of Prostaglandins (PG) and Thromboxanes (TX) / prostaglandin-endoperoxide synthase / prostaglandin-endoperoxide synthase activity / cyclooxygenase pathway / negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress / positive regulation of prostaglandin biosynthetic process / regulation of neuroinflammatory response / positive regulation of fever generation / response to selenium ion / prostaglandin secretion / cellular response to fluid shear stress / response to nematode / nuclear inner membrane / prostaglandin biosynthetic process / dioxygenase activity / bone mineralization / nuclear outer membrane / decidualization / brown fat cell differentiation / keratinocyte differentiation / positive regulation of brown fat cell differentiation / embryo implantation / peroxidase activity / regulation of blood pressure / regulation of cell population proliferation / response to oxidative stress / cellular response to hypoxia / neuron projection / heme binding / endoplasmic reticulum membrane / negative regulation of apoptotic process / enzyme binding / protein homodimerization activity / metal ion binding / cytosol / cytoplasm Similarity search - Function | ||||||
| Biological species |  | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.9 Å MOLECULAR REPLACEMENT / Resolution: 2.9 Å | ||||||
 Authors Authors | Kurumbail, R. / Stallings, W. | ||||||
 Citation Citation |  Journal: Nature / Year: 1996 Journal: Nature / Year: 1996Title: Structural basis for selective inhibition of cyclooxygenase-2 by anti-inflammatory agents. Authors: Kurumbail, R.G. / Stevens, A.M. / Gierse, J.K. / McDonald, J.J. / Stegeman, R.A. / Pak, J.Y. / Gildehaus, D. / Miyashiro, J.M. / Penning, T.D. / Seibert, K. / Isakson, P.C. / Stallings, W.C. #1:  Journal: Nature / Year: 1997 Journal: Nature / Year: 1997Title: Erratum. Structural Basis for Selective Inhibition of Cyclooxygenase-2 by Anti-Inflammatory Agents Authors: Kurumbail, R.G. / Stevens, A.M. / Gierse, J.K. / Mcdonald, J.J. / Stegeman, R.A. / Pak, J.Y. / Gildehaus, D. / Miyashiro, J.M. / Penning, T.D. / Seibert, K. / Isakson, P.C. / Stallings, W.C. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4cox.cif.gz 4cox.cif.gz | 444.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4cox.ent.gz pdb4cox.ent.gz | 363 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4cox.json.gz 4cox.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Summary document |  4cox_validation.pdf.gz 4cox_validation.pdf.gz | 2.8 MB | Display |  wwPDB validaton report wwPDB validaton report |
|---|---|---|---|---|
| Full document |  4cox_full_validation.pdf.gz 4cox_full_validation.pdf.gz | 2.9 MB | Display | |
| Data in XML |  4cox_validation.xml.gz 4cox_validation.xml.gz | 90.8 KB | Display | |
| Data in CIF |  4cox_validation.cif.gz 4cox_validation.cif.gz | 119.3 KB | Display | |
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/co/4cox https://data.pdbj.org/pub/pdb/validation_reports/co/4cox ftp://data.pdbj.org/pub/pdb/validation_reports/co/4cox ftp://data.pdbj.org/pub/pdb/validation_reports/co/4cox | HTTPS FTP |
-Related structure data
| Related structure data |  1cx2C  3pghC  5coxC  6coxC  1prhS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||||||||||||||||||||||||
| 2 | 
| ||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS oper:
|
- Components
Components
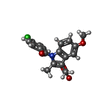
| #1: Protein | Mass: 67318.719 Da / Num. of mol.: 4 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   References: UniProt: Q05769, prostaglandin-endoperoxide synthase #2: Sugar | ChemComp-NAG / #3: Chemical | ChemComp-HEM / #4: Chemical | ChemComp-IMN / Has protein modification | Y | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.54 Å3/Da / Density % sol: 52 % / Description: PGHS-1 DIMER WAS USED AS THE SEARCH MODEL. | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Crystal grow | pH: 8 Details: 20 MM SODIUM PHOSPHATE, 100 MM NACL, 0.6% BETA- OCTYLGLUCOSIDE, 10 MG/ML PROTEIN, 1MM INHIBITOR MIXED WITH A RESERVOIR SOLUTION CONTAINING 20-34% MONOMETHYL PEG 550, 10-240 MGCL2, 50 MM EPPS ...Details: 20 MM SODIUM PHOSPHATE, 100 MM NACL, 0.6% BETA- OCTYLGLUCOSIDE, 10 MG/ML PROTEIN, 1MM INHIBITOR MIXED WITH A RESERVOIR SOLUTION CONTAINING 20-34% MONOMETHYL PEG 550, 10-240 MGCL2, 50 MM EPPS PH 8.0 IN THE RATIO OF 1:1 | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 18 ℃ / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 113 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 |
| Detector | Type: MAR scanner 300 mm plate / Detector: IMAGE PLATE / Date: Aug 1, 1995 / Details: SUPPER MIRRORS |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength: 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.9→40 Å / Num. obs: 50058 / % possible obs: 77.9 % / Observed criterion σ(I): -1.5 / Redundancy: 3.3 % / Rmerge(I) obs: 0.135 / Rsym value: 0.135 / Net I/σ(I): 8 |
| Reflection shell | Resolution: 2.9→3 Å / Redundancy: 2.6 % / Rmerge(I) obs: 0.488 / Mean I/σ(I) obs: 1.77 / Rsym value: 0.488 / % possible all: 67.5 |
| Reflection | *PLUS Num. measured all: 1145408 |
| Reflection shell | *PLUS % possible obs: 67.5 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure:  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PROSTAGLANDIN SYNTHASE-1, PDB ENTRY 1PRH Resolution: 2.9→8 Å / Data cutoff high absF: 100000 / Data cutoff low absF: 0.1 / σ(F): 2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 12 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.9→8 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.9→3.03 Å / Total num. of bins used: 8
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.1F / Classification: refinement X-PLOR / Version: 3.1F / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor obs: 0.264 |
 Movie
Movie Controller
Controller

















 PDBj
PDBj