+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
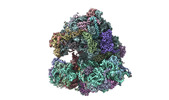
| タイトル | High-resolution C. elegans 80S ribosome structure - class 1 | |||||||||
 マップデータ マップデータ | Sharpened map | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | ribosome / endogenous purified 80S ribosome | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報Translesion synthesis by REV1 / Recognition of DNA damage by PCNA-containing replication complex / Translesion Synthesis by POLH / mTORC1-mediated signalling / SRP-dependent cotranslational protein targeting to membrane / SCF(Skp2)-mediated degradation of p27/p21 / Downregulation of TGF-beta receptor signaling / TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) / SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription / Regulation of PLK1 Activity at G2/M Transition ...Translesion synthesis by REV1 / Recognition of DNA damage by PCNA-containing replication complex / Translesion Synthesis by POLH / mTORC1-mediated signalling / SRP-dependent cotranslational protein targeting to membrane / SCF(Skp2)-mediated degradation of p27/p21 / Downregulation of TGF-beta receptor signaling / TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) / SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription / Regulation of PLK1 Activity at G2/M Transition / Regulation of innate immune responses to cytosolic DNA / Autodegradation of the E3 ubiquitin ligase COP1 / ABC-family proteins mediated transport / Degradation of DVL / Regulation of FZD by ubiquitination / N-glycan trimming in the ER and Calnexin/Calreticulin cycle / Hedgehog ligand biogenesis / Hedgehog 'on' state / Translesion synthesis by POLK / Translesion synthesis by POLI / Negative regulation of MAPK pathway / MAPK6/MAPK4 signaling / UCH proteinases / Josephin domain DUBs / DNA Damage Recognition in GG-NER / Gap-filling DNA repair synthesis and ligation in GG-NER / Dual Incision in GG-NER / Major pathway of rRNA processing in the nucleolus and cytosol / CDK-mediated phosphorylation and removal of Cdc6 / Cyclin D associated events in G1 / Ubiquitin Mediated Degradation of Phosphorylated Cdc25A / Formation of a pool of free 40S subunits / Formation of the ternary complex, and subsequently, the 43S complex / GTP hydrolysis and joining of the 60S ribosomal subunit / Ubiquitin-dependent degradation of Cyclin D / PTK6 Regulates RTKs and Their Effectors AKT1 and DOK1 / FBXL7 down-regulates AURKA during mitotic entry and in early mitosis / Regulation of RUNX2 expression and activity / Regulation of RUNX3 expression and activity / Regulation of PTEN stability and activity / ER Quality Control Compartment (ERQC) / Interleukin-1 signaling / Endosomal Sorting Complex Required For Transport (ESCRT) / Protein hydroxylation / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / Oxygen-dependent proline hydroxylation of Hypoxia-inducible Factor Alpha / Downregulation of ERBB4 signaling / Formation of Incision Complex in GG-NER / E3 ubiquitin ligases ubiquitinate target proteins / Regulation of PTEN localization / Aggrephagy / GSK3B and BTRC:CUL1-mediated-degradation of NFE2L2 / L13a-mediated translational silencing of Ceruloplasmin expression / Stimuli-sensing channels / Orc1 removal from chromatin / Translation initiation complex formation / Ribosomal scanning and start codon recognition / Neddylation / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Metalloprotease DUBs / Downregulation of ERBB2 signaling / Negative regulators of DDX58/IFIH1 signaling / RAS processing / Degradation of beta-catenin by the destruction complex / Regulation of TNFR1 signaling / Formation of TC-NER Pre-Incision Complex / Dual incision in TC-NER / Gap-filling DNA repair synthesis and ligation in TC-NER / Synthesis of active ubiquitin: roles of E1 and E2 enzymes / Iron uptake and transport / KEAP1-NFE2L2 pathway / Antigen processing: Ubiquitination & Proteasome degradation / Downregulation of SMAD2/3:SMAD4 transcriptional activity / Ovarian tumor domain proteases / Negative regulation of MET activity / EGFR downregulation / Cargo recognition for clathrin-mediated endocytosis / Peroxisomal protein import / Regulation of signaling by CBL / Ub-specific processing proteases / Clathrin-mediated endocytosis / polarity specification of anterior/posterior axis / cytoplasmic side of rough endoplasmic reticulum membrane / endonucleolytic cleavage to generate mature 3'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / protein-RNA complex assembly / rough endoplasmic reticulum / endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of LSU-rRNA / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / ribosomal large subunit biogenesis / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / maturation of SSU-rRNA / determination of adult lifespan / small-subunit processome / positive regulation of apoptotic signaling pathway / maintenance of translational fidelity / modification-dependent protein catabolic process / protein tag activity / rRNA processing / ribosomal small subunit biogenesis 類似検索 - 分子機能 | |||||||||
| 生物種 |  | |||||||||
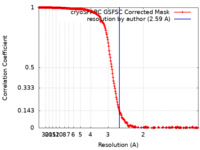
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.59 Å | |||||||||
 データ登録者 データ登録者 | Sehgal E / Serrao VHB / Arribere J | |||||||||
| 資金援助 |  米国, 1件 米国, 1件
| |||||||||
 引用 引用 |  ジャーナル: RNA / 年: 2024 ジャーナル: RNA / 年: 2024タイトル: High-resolution reconstruction of a ribosome sheds light on evolutionary dynamics and tissue specificity. 著者: Enisha Sehgal / Chloe Wohlenberg / Evan M Soukup / Marcus J Viscardi / Vitor Hugo Balasco Serrão / Joshua A Arribere /  要旨: is an important model organism for human health and disease, with foundational contributions to the understanding of gene expression and tissue patterning in animals. An invaluable tool in modern ... is an important model organism for human health and disease, with foundational contributions to the understanding of gene expression and tissue patterning in animals. An invaluable tool in modern gene expression research is the presence of a high-resolution ribosome structure, though no such structure exists for Here, we present a high-resolution single-particle cryogenic electron microscopy (cryo-EM) reconstruction and molecular model of a ribosome, revealing a significantly streamlined animal ribosome. Many facets of ribosome structure are conserved in , including overall ribosomal architecture and the mechanism of cycloheximide, whereas other facets, such as expansion segments and eL28, are rapidly evolving. We identify uL5 and uL23 as two instances of tissue-specific ribosomal protein paralog expression conserved in , suggesting that ribosomes vary across tissues. The ribosome structure will provide a basis for future structural, biochemical, and genetic studies of translation in this important animal system. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_45392.map.gz emd_45392.map.gz | 1.1 GB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-45392-v30.xml emd-45392-v30.xml emd-45392.xml emd-45392.xml | 103 KB 103 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_45392_fsc.xml emd_45392_fsc.xml | 22.6 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_45392.png emd_45392.png | 78.3 KB | ||
| Filedesc metadata |  emd-45392.cif.gz emd-45392.cif.gz | 19.1 KB | ||
| その他 |  emd_45392_additional_1.map.gz emd_45392_additional_1.map.gz emd_45392_half_map_1.map.gz emd_45392_half_map_1.map.gz emd_45392_half_map_2.map.gz emd_45392_half_map_2.map.gz | 597.2 MB 1.1 GB 1.1 GB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-45392 http://ftp.pdbj.org/pub/emdb/structures/EMD-45392 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45392 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-45392 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_45392_validation.pdf.gz emd_45392_validation.pdf.gz | 1.3 MB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_45392_full_validation.pdf.gz emd_45392_full_validation.pdf.gz | 1.3 MB | 表示 | |
| XML形式データ |  emd_45392_validation.xml.gz emd_45392_validation.xml.gz | 32.5 KB | 表示 | |
| CIF形式データ |  emd_45392_validation.cif.gz emd_45392_validation.cif.gz | 43.2 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45392 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45392 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45392 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-45392 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  9caiMC  9bh5C C: 同じ文献を引用 ( M: このマップから作成された原子モデル |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_45392.map.gz / 形式: CCP4 / 大きさ: 1.2 GB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_45392.map.gz / 形式: CCP4 / 大きさ: 1.2 GB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | Sharpened map | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.65 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
- 試料の構成要素
試料の構成要素
+全体 : Caenorhabditis elegans 80S ribosome
+超分子 #1: Caenorhabditis elegans 80S ribosome
+分子 #1: Small ribosomal subunit protein uS2
+分子 #2: Large ribosomal subunit protein uL2
+分子 #4: Small ribosomal subunit protein eS1
+分子 #5: Large ribosomal subunit protein uL3
+分子 #6: Small ribosomal subunit protein uS5
+分子 #7: Small ribosomal subunit protein uS3
+分子 #8: Small ribosomal subunit protein eS4
+分子 #13: Small ribosomal subunit protein eS17
+分子 #14: Plectin/S10 N-terminal domain-containing protein
+分子 #15: Ribosomal Protein, Small subunit
+分子 #16: Small ribosomal subunit protein uS13
+分子 #17: Small ribosomal subunit protein eS19
+分子 #18: Small ribosomal subunit protein eS26
+分子 #19: Ubiquitin-like protein 1-ribosomal protein eS31 fusion protein
+分子 #20: Small ribosomal subunit protein uS11
+分子 #21: Small ribosomal subunit protein eS8
+分子 #22: Small ribosomal subunit protein uS7
+分子 #23: Small ribosomal subunit protein eS12
+分子 #24: Small ribosomal subunit protein eS21
+分子 #25: Small ribosomal subunit protein uS15
+分子 #26: Small ribosomal subunit protein eS25
+分子 #27: Small ribosomal subunit protein uS14
+分子 #28: Small ribosomal subunit protein eS24
+分子 #29: Small ribosomal subunit protein uS10
+分子 #30: Small ribosomal subunit protein eS6
+分子 #31: Small ribosomal subunit protein eS27
+分子 #32: Small ribosomal subunit protein uS19
+分子 #33: Small ribosomal subunit protein eS28
+分子 #34: Ubiquitin-like domain-containing protein
+分子 #35: Small ribosomal subunit protein uS12
+分子 #36: Small ribosomal subunit protein uS17
+分子 #37: Small ribosomal subunit protein uS4
+分子 #38: Small ribosomal subunit protein uS9
+分子 #39: Small ribosomal subunit protein eS7
+分子 #40: Large ribosomal subunit protein uL30
+分子 #41: Large ribosomal subunit protein eL20
+分子 #42: Large ribosomal subunit protein uL14
+分子 #43: Large ribosomal subunit protein eL22
+分子 #44: Large ribosomal subunit protein eL13
+分子 #45: Large ribosomal subunit protein eL8
+分子 #46: Large ribosomal subunit protein uL16
+分子 #47: Large ribosomal subunit protein uL13
+分子 #48: Large ribosomal subunit protein uL4
+分子 #49: Large ribosomal subunit protein uL29
+分子 #50: Large ribosomal subunit protein eL6
+分子 #51: Large ribosomal subunit protein eL42
+分子 #52: Large ribosomal subunit protein eL33
+分子 #53: Large ribosomal subunit protein eL36
+分子 #54: Large ribosomal subunit protein eL39
+分子 #55: Large ribosomal subunit protein eL15
+分子 #56: Large ribosomal subunit protein uL15
+分子 #57: Large ribosomal subunit protein eL43
+分子 #58: Large ribosomal subunit protein eL14
+分子 #59: Large ribosomal subunit protein eL30
+分子 #60: Large ribosomal subunit protein uL6
+分子 #61: Large ribosomal subunit protein uL24
+分子 #62: Large ribosomal subunit protein eL19
+分子 #63: 60S ribosomal protein L29
+分子 #64: Large ribosomal subunit protein eL18
+分子 #65: Large ribosomal subunit protein eL21
+分子 #66: Large ribosomal subunit protein uL18
+分子 #67: Large ribosomal subunit protein eL27
+分子 #68: Large ribosomal subunit protein uL22
+分子 #69: eL41
+分子 #70: Large ribosomal subunit protein eL34
+分子 #71: Large ribosomal subunit protein eL38
+分子 #72: Ubiquitin-ribosomal protein eL40 fusion protein
+分子 #73: Large ribosomal subunit protein eL31
+分子 #74: Large ribosomal subunit protein eL32
+分子 #75: Large ribosomal subunit protein eL24
+分子 #76: Ribosomal protein L37
+分子 #77: Large ribosomal subunit protein uL5B
+分子 #78: Large ribosomal subunit protein uL23B
+分子 #3: tRNA
+分子 #9: 5S rRNA
+分子 #10: 5.8S rRNA
+分子 #11: 18S rRNA
+分子 #12: 28S rRNA
+分子 #79: mRNA
+分子 #80: SPERMIDINE
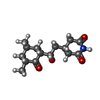
+分子 #81: 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethy...
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 0.5 mg/mL |
|---|---|
| 緩衝液 | pH: 7.4 |
| グリッド | モデル: Quantifoil R2/1 / 材質: COPPER / メッシュ: 200 / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: CONTINUOUS / 支持フィルム - Film thickness: 2 / 前処理 - タイプ: GLOW DISCHARGE / 前処理 - 時間: 20 sec. |
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 298 K / 装置: FEI VITROBOT MARK IV |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | TFS KRIOS |
|---|---|
| ソフトウェア | 名称: SerialEM (ver. v4.93) |
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOCONTINUUM (6k x 4k) 撮影したグリッド数: 1 / 実像数: 8337 / 平均電子線量: 40.94 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.0 µm / 最小 デフォーカス(公称値): 0.5 µm |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
+ 画像解析
画像解析
-原子モデル構築 1
| 初期モデル | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| ソフトウェア | 名称:  Coot Coot |
| 精密化 | 空間: REAL / プロトコル: RIGID BODY FIT / 温度因子: 59.4 |
| 得られたモデル |  PDB-9cai: |
 ムービー
ムービー コントローラー
コントローラー












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)