[English] 日本語
 Yorodumi
Yorodumi- EMDB-44129: Open state of kainate receptor GluK2 in complex with agonist glut... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Open state of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to two concanavalin A dimers. Composite map. | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | kainate receptor / GluK2 / positive allosteric modulator / BPAM344 / open / concanavalin A / ConA / glutamate / MEMBRANE PROTEIN | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationmossy fiber rosette / detection of cold stimulus involved in thermoception / Activation of Na-permeable kainate receptors / kainate selective glutamate receptor complex / Activation of Ca-permeable Kainate Receptor / D-glucose binding / negative regulation of synaptic transmission, glutamatergic / regulation of short-term neuronal synaptic plasticity / inhibitory postsynaptic potential / ubiquitin conjugating enzyme binding ...mossy fiber rosette / detection of cold stimulus involved in thermoception / Activation of Na-permeable kainate receptors / kainate selective glutamate receptor complex / Activation of Ca-permeable Kainate Receptor / D-glucose binding / negative regulation of synaptic transmission, glutamatergic / regulation of short-term neuronal synaptic plasticity / inhibitory postsynaptic potential / ubiquitin conjugating enzyme binding / glutamate receptor activity / receptor clustering / modulation of excitatory postsynaptic potential / extracellularly glutamate-gated ion channel activity / D-mannose binding / regulation of JNK cascade / kainate selective glutamate receptor activity / ionotropic glutamate receptor complex / behavioral fear response / neuronal action potential / positive regulation of synaptic transmission / glutamate-gated receptor activity / glutamate-gated calcium ion channel activity / ligand-gated monoatomic ion channel activity involved in regulation of presynaptic membrane potential / presynaptic modulation of chemical synaptic transmission / dendrite cytoplasm / hippocampal mossy fiber to CA3 synapse / regulation of membrane potential / SNARE binding / excitatory postsynaptic potential / synaptic transmission, glutamatergic / transmitter-gated monoatomic ion channel activity involved in regulation of postsynaptic membrane potential / PDZ domain binding / postsynaptic density membrane / regulation of long-term neuronal synaptic plasticity / modulation of chemical synaptic transmission / terminal bouton / intracellular calcium ion homeostasis / vasodilation / positive regulation of neuron apoptotic process / manganese ion binding / presynaptic membrane / scaffold protein binding / chemical synaptic transmission / perikaryon / postsynaptic membrane / neuron apoptotic process / negative regulation of neuron apoptotic process / postsynaptic density / axon / neuronal cell body / glutamatergic synapse / ubiquitin protein ligase binding / dendrite / calcium ion binding / synapse / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |   | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 4.29 Å | |||||||||||||||
 Authors Authors | Nadezhdin KD / Gangwar SP / Sobolevsky AI | |||||||||||||||
| Funding support |  United States, 4 items United States, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Nature / Year: 2024 Journal: Nature / Year: 2024Title: Kainate receptor channel opening and gating mechanism. Authors: Shanti Pal Gangwar / Maria V Yelshanskaya / Kirill D Nadezhdin / Laura Y Yen / Thomas P Newton / Muhammed Aktolun / Maria G Kurnikova / Alexander I Sobolevsky /  Abstract: Kainate receptors, a subclass of ionotropic glutamate receptors, are tetrameric ligand-gated ion channels that mediate excitatory neurotransmission. Kainate receptors modulate neuronal circuits and ...Kainate receptors, a subclass of ionotropic glutamate receptors, are tetrameric ligand-gated ion channels that mediate excitatory neurotransmission. Kainate receptors modulate neuronal circuits and synaptic plasticity during the development and function of the central nervous system and are implicated in various neurological and psychiatric diseases, including epilepsy, depression, schizophrenia, anxiety and autism. Although structures of kainate receptor domains and subunit assemblies are available, the mechanism of kainate receptor gating remains poorly understood. Here we present cryo-electron microscopy structures of the kainate receptor GluK2 in the presence of the agonist glutamate and the positive allosteric modulators lectin concanavalin A and BPAM344. Concanavalin A and BPAM344 inhibit kainate receptor desensitization and prolong activation by acting as a spacer between the amino-terminal and ligand-binding domains and a stabilizer of the ligand-binding domain dimer interface, respectively. Channel opening involves the kinking of all four pore-forming M3 helices. Our structures reveal the molecular basis of kainate receptor gating, which could guide the development of drugs for treatment of neurological disorders. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44129.map.gz emd_44129.map.gz | 55.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44129-v30.xml emd-44129-v30.xml emd-44129.xml emd-44129.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_44129.png emd_44129.png | 122.7 KB | ||
| Filedesc metadata |  emd-44129.cif.gz emd-44129.cif.gz | 7.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44129 http://ftp.pdbj.org/pub/emdb/structures/EMD-44129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44129 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44129 | HTTPS FTP |
-Validation report
| Summary document |  emd_44129_validation.pdf.gz emd_44129_validation.pdf.gz | 501.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_44129_full_validation.pdf.gz emd_44129_full_validation.pdf.gz | 501.4 KB | Display | |
| Data in XML |  emd_44129_validation.xml.gz emd_44129_validation.xml.gz | 6.5 KB | Display | |
| Data in CIF |  emd_44129_validation.cif.gz emd_44129_validation.cif.gz | 7.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44129 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44129 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44129 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44129 | HTTPS FTP |
-Related structure data
| Related structure data |  9b36MC  9b33C  9b34C  9b35C  9b37C  9b38C  9b39C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_44129.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44129.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3487 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
-Entire : full-length rat GluK2 tetramer in complex with two concanavalin A...
| Entire | Name: full-length rat GluK2 tetramer in complex with two concanavalin A dimers |
|---|---|
| Components |
|
-Supramolecule #1: full-length rat GluK2 tetramer in complex with two concanavalin A...
| Supramolecule | Name: full-length rat GluK2 tetramer in complex with two concanavalin A dimers type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 510 KDa |
-Macromolecule #1: Glutamate receptor ionotropic, kainate 2
| Macromolecule | Name: Glutamate receptor ionotropic, kainate 2 / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 102.976586 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MKIISPVLSN LVFSRSIKVL LCLLWIGYSQ GTTHVLRFGG IFEYVESGPM GAEELAFRFA VNTINRNRTL LPNTTLTYDT QKINLYDSF EASKKACDQL SLGVAAIFGP SHSSSANAVQ SICNALGVPH IQTRWKHQVS DNKDSFYVSL YPDFSSLSRA I LDLVQFFK ...String: MKIISPVLSN LVFSRSIKVL LCLLWIGYSQ GTTHVLRFGG IFEYVESGPM GAEELAFRFA VNTINRNRTL LPNTTLTYDT QKINLYDSF EASKKACDQL SLGVAAIFGP SHSSSANAVQ SICNALGVPH IQTRWKHQVS DNKDSFYVSL YPDFSSLSRA I LDLVQFFK WKTVTVVYDD STGLIRLQEL IKAPSRYNLR LKIRQLPADT KDAKPLLKEM KRGKEFHVIF DCSHEMAAGI LK QALAMGM MTEYYHYIFT TLDLFALDVE PYRYSGVNMT GFRILNTENT QVSSIIEKWS MERLQAPPKP DSGLLDGFMT TDA ALMYDA VHVVSVAVQQ FPQMTVSSLQ CNRHKPWRFG TRFMSLIKEA HWEGLTGRIT FNKTNGLRTD FDLDVISLKE EGLE KIGTW DPASGLNMTE SQKGKPANIT DSLSNRSLIV TTILEEPYVL FKKSDKPLYG NDRFEGYCID LLRELSTILG FTYEI RLVE DGKYGAQDDV NGQWNGMVRE LIDHKADLAV APLAITYVRE KVIDFSKPFM TLGISILYRK PNGTNPGVFS FLNPLS PDI WMYVLLACLG VSCVLFVIAR FSPYEWYNPH PCNPDSDVVE NNFTLLNSFW FGVGALMQQG SELMPKALST RIVGGIW WF FTLIIISSYT ANLAAFLTVE RMESPIDSAD DLAKQTKIEY GAVEDGATMT FFKKSKISTY DKMWAFMSSR RQSVLVKS N EEGIQRVLTS DYAFLMESTT IEFVTQRNCN LTQIGGLIDS KGYGVGTPMG SPYRDKITIA ILQLQEEGKL HMMKEKWWR GNGCPEEESK EASALGVQNI GGIFIVLAAG LVLSVFVAVG EFLYKSKKNA QLEKRSFCSA MVEELRMSLK CQRRLKHKPQ APVIVKTEE VINMHTFNDR RLPGKETMAL VPR UniProtKB: Glutamate receptor ionotropic, kainate 2 |
-Macromolecule #2: Concanavalin A
| Macromolecule | Name: Concanavalin A / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 25.622385 KDa |
| Sequence | String: ADTIVAVELD TYPNTDIGDP SYPHIGIDIK SVRSKKTAKW NMQNGKVGTA HIIYNSVDKR LSAVVSYPNA DSATVSYDVD LDNVLPEWV RVGLSASTGL YKETNTILSW SFTSKLKSNS THETNALHFM FNQFSKDQKD LILQGDATTG TDGNLELTRV S SNGSPQGS ...String: ADTIVAVELD TYPNTDIGDP SYPHIGIDIK SVRSKKTAKW NMQNGKVGTA HIIYNSVDKR LSAVVSYPNA DSATVSYDVD LDNVLPEWV RVGLSASTGL YKETNTILSW SFTSKLKSNS THETNALHFM FNQFSKDQKD LILQGDATTG TDGNLELTRV S SNGSPQGS SVGRALFYAP VHIWESSAVV ASFEATFTFL IKSPDSHPAD GIAFFISNID SSIPSGSTGR LLGLFPDAN UniProtKB: Concanavalin V |
-Macromolecule #10: 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-...
| Macromolecule | Name: 4-cyclopropyl-7-fluoro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide type: ligand / ID: 10 / Number of copies: 4 / Formula: 2J9 |
|---|---|
| Molecular weight | Theoretical: 242.27 Da |
| Chemical component information | 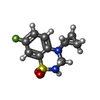 ChemComp-2J9: |
-Macromolecule #11: GLUTAMIC ACID
| Macromolecule | Name: GLUTAMIC ACID / type: ligand / ID: 11 / Number of copies: 4 / Formula: GLU |
|---|---|
| Molecular weight | Theoretical: 147.129 Da |
| Chemical component information |  ChemComp-GLU: |
-Macromolecule #12: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(tri...
| Macromolecule | Name: (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate type: ligand / ID: 12 / Number of copies: 14 / Formula: POV |
|---|---|
| Molecular weight | Theoretical: 760.076 Da |
| Chemical component information |  ChemComp-POV: |
-Macromolecule #13: CHOLESTEROL
| Macromolecule | Name: CHOLESTEROL / type: ligand / ID: 13 / Number of copies: 8 / Formula: CLR |
|---|---|
| Molecular weight | Theoretical: 386.654 Da |
| Chemical component information |  ChemComp-CLR: |
-Macromolecule #14: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 14 / Number of copies: 4 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #15: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 15 / Number of copies: 4 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Component:
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Support film - Material: GOLD / Support film - topology: HOLEY | ||||||||||||||||||||||||
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Digitization - Dimensions - Width: 5760 pixel / Digitization - Dimensions - Height: 4092 pixel / Number grids imaged: 5 / Number real images: 22990 / Average exposure time: 2.5 sec. / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-9b36: |
 Movie
Movie Controller
Controller
















 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)




















