+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of human HGSNAT bound with Acetyl-CoA | |||||||||
 Map data Map data | final sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Membrane protein / Lysosome / Transmembrane acetyltransferase / Acetyl-CoA / Heperan sulfate | |||||||||
| Function / homology | :  Function and homology information Function and homology information | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.92 Å | |||||||||
 Authors Authors | Li F / Zhao B | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Structural and mechanistic insights into a lysosomal membrane enzyme HGSNAT involved in Sanfilippo syndrome. Authors: Boyang Zhao / Zhongzheng Cao / Yi Zheng / Phuong Nguyen / Alisa Bowen / Robert H Edwards / Robert M Stroud / Yi Zhou / Menno Van Lookeren Campagne / Fei Li /  Abstract: Heparan sulfate (HS) is degraded in lysosome by a series of glycosidases. Before the glycosidases can act, the terminal glucosamine of HS must be acetylated by the integral lysosomal membrane enzyme ...Heparan sulfate (HS) is degraded in lysosome by a series of glycosidases. Before the glycosidases can act, the terminal glucosamine of HS must be acetylated by the integral lysosomal membrane enzyme heparan-α-glucosaminide N-acetyltransferase (HGSNAT). Mutations of HGSNAT cause HS accumulation and consequently mucopolysaccharidosis IIIC, a devastating lysosomal storage disease characterized by progressive neurological deterioration and early death where no treatment is available. HGSNAT catalyzes a unique transmembrane acetylation reaction where the acetyl group of cytosolic acetyl-CoA is transported across the lysosomal membrane and attached to HS in one reaction. However, the reaction mechanism remains elusive. Here we report six cryo-EM structures of HGSNAT along the reaction pathway. These structures reveal a dimer arrangement and a unique structural fold, which enables the elucidation of the reaction mechanism. We find that a central pore within each monomer traverses the membrane and controls access of cytosolic acetyl-CoA to the active site at its luminal mouth where glucosamine binds. A histidine-aspartic acid catalytic dyad catalyzes the transfer reaction via a ternary complex mechanism. Furthermore, the structures allow the mapping of disease-causing variants and reveal their potential impact on the function, thus creating a framework to guide structure-based drug discovery efforts. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43319.map.gz emd_43319.map.gz | 307 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43319-v30.xml emd-43319-v30.xml emd-43319.xml emd-43319.xml | 17.2 KB 17.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43319_fsc.xml emd_43319_fsc.xml | 14.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_43319.png emd_43319.png | 96.7 KB | ||
| Filedesc metadata |  emd-43319.cif.gz emd-43319.cif.gz | 6.1 KB | ||
| Others |  emd_43319_additional_1.map.gz emd_43319_additional_1.map.gz emd_43319_half_map_1.map.gz emd_43319_half_map_1.map.gz emd_43319_half_map_2.map.gz emd_43319_half_map_2.map.gz | 162.5 MB 302 MB 302 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43319 http://ftp.pdbj.org/pub/emdb/structures/EMD-43319 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43319 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43319 | HTTPS FTP |
-Validation report
| Summary document |  emd_43319_validation.pdf.gz emd_43319_validation.pdf.gz | 916.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43319_full_validation.pdf.gz emd_43319_full_validation.pdf.gz | 916.4 KB | Display | |
| Data in XML |  emd_43319_validation.xml.gz emd_43319_validation.xml.gz | 23.9 KB | Display | |
| Data in CIF |  emd_43319_validation.cif.gz emd_43319_validation.cif.gz | 30.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43319 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43319 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43319 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43319 | HTTPS FTP |
-Related structure data
| Related structure data |  8vkjMC  8vlgC  8vliC  8vluC  8vlvC  8vlyC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_43319.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43319.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | final sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.813 Å | ||||||||||||||||||||||||||||||||||||
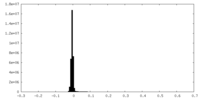
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: unsharpened map
| File | emd_43319_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | unsharpened map | ||||||||||||
| Projections & Slices |
| ||||||||||||
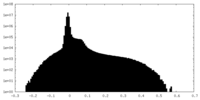
| Density Histograms |
-Half map: half map B
| File | emd_43319_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
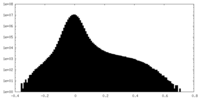
| Density Histograms |
-Half map: half map A
| File | emd_43319_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
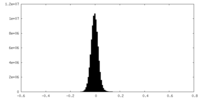
| Density Histograms |
- Sample components
Sample components
-Entire : Purified human HGSNAT
| Entire | Name: Purified human HGSNAT |
|---|---|
| Components |
|
-Supramolecule #1: Purified human HGSNAT
| Supramolecule | Name: Purified human HGSNAT / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 73 KDa |
-Macromolecule #1: Heparan-alpha-glucosaminide N-acetyltransferase
| Macromolecule | Name: Heparan-alpha-glucosaminide N-acetyltransferase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: ec: 2.3.1.78 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 73.365039 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MTGARASAAE QRRAGRSGQA RAAERAAGMS GAGRALAALL LAASVLSAAL LAPGGSSGRD AQAAPPRDLD KKRHAELKMD QALLLIHNE LLWTNLTVYW KSECCYHCLF QVLVNVPQSP KAGKPSAAAA SVSTQHGSIL QLNDTLEEKE VCRLEYRFGE F GNYSLLVK ...String: MTGARASAAE QRRAGRSGQA RAAERAAGMS GAGRALAALL LAASVLSAAL LAPGGSSGRD AQAAPPRDLD KKRHAELKMD QALLLIHNE LLWTNLTVYW KSECCYHCLF QVLVNVPQSP KAGKPSAAAA SVSTQHGSIL QLNDTLEEKE VCRLEYRFGE F GNYSLLVK NIHNGVSEIA CDLAVNEDPV DSNLPVSIAF LIGLAVIIVI SFLRLLLSLD DFNNWISKAI SSRETDRLIN SE LGSPSRT DPLDGDVQPA TWRLSALPPR LRSVDTFRGI ALILMVFVNY GGGKYWYFKH ASWNGLTVAD LVFPWFVFIM GSS IFLSMT SILQRGCSKF RLLGKIAWRS FLLICIGIII VNPNYCLGPL SWDKVRIPGV LQRLGVTYFV VAVLELLFAK PVPE HCASE RSCLSLRDIT SSWPQWLLIL VLEGLWLGLT FLLPVPGCPT GYLGPGGIGD FGKYPNCTGG AAGYIDRLLL GDDHL YQHP SSAVLYHTEV AYDPEGILGT INSIVMAFLG VQAGKILLYY KARTKDILIR FTAWCCILGL ISVALTKVSE NEGFIP VNK NLWSLSYVTT LSSFAFFILL VLYPVVDVKG LWTGTPFFYP GMNSILVYVG HEVFENYFPF QWKLKDNQSH KEHLTQN IV ATALWVLIAY ILYRKKIFWK I UniProtKB: UNIPROTKB: Q68CP4 |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 5 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Macromolecule #4: ACETYL COENZYME *A
| Macromolecule | Name: ACETYL COENZYME *A / type: ligand / ID: 4 / Number of copies: 2 / Formula: ACO |
|---|---|
| Molecular weight | Theoretical: 809.571 Da |
| Chemical component information |  ChemComp-ACO: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 3.1 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 1.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 22.0 µm / Nominal defocus min: 10.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)