[English] 日本語
 Yorodumi
Yorodumi- EMDB-42435: Major interface of Streptococcal surface enolase dimer from AP53 ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Major interface of Streptococcal surface enolase dimer from AP53 group A streptococcus bound to a lipid vesicle | |||||||||
 Map data Map data | SEN dimer map on the surface of lipid major dimer interface | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Glycolysis / plasminogen binder / membrane protein / LYASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationphosphopyruvate hydratase / phosphopyruvate hydratase complex / phosphopyruvate hydratase activity / peptidoglycan-based cell wall / glycolytic process / cell surface / magnesium ion binding / extracellular region Similarity search - Function | |||||||||
| Biological species |  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.8 Å | |||||||||
 Authors Authors | Tjia-Fleck S / Readnour BM / Castellino FJ | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Streptococcus surface alpha enolase exposed dimers were found to be the active form on lipid surface that binds to human plasminogen Authors: Tjia-Fleck S / Readnour BM / Castellino FJ | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42435.map.gz emd_42435.map.gz | 35.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42435-v30.xml emd-42435-v30.xml emd-42435.xml emd-42435.xml | 17.8 KB 17.8 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42435_fsc.xml emd_42435_fsc.xml | 7.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_42435.png emd_42435.png | 92.8 KB | ||
| Filedesc metadata |  emd-42435.cif.gz emd-42435.cif.gz | 5.8 KB | ||
| Others |  emd_42435_additional_1.map.gz emd_42435_additional_1.map.gz emd_42435_half_map_1.map.gz emd_42435_half_map_1.map.gz emd_42435_half_map_2.map.gz emd_42435_half_map_2.map.gz | 35.2 MB 34.7 MB 34.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42435 http://ftp.pdbj.org/pub/emdb/structures/EMD-42435 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42435 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42435 | HTTPS FTP |
-Validation report
| Summary document |  emd_42435_validation.pdf.gz emd_42435_validation.pdf.gz | 726.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42435_full_validation.pdf.gz emd_42435_full_validation.pdf.gz | 726.1 KB | Display | |
| Data in XML |  emd_42435_validation.xml.gz emd_42435_validation.xml.gz | 14.4 KB | Display | |
| Data in CIF |  emd_42435_validation.cif.gz emd_42435_validation.cif.gz | 18.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42435 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42435 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42435 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42435 | HTTPS FTP |
-Related structure data
| Related structure data |  8uopMC  8uoyC  8uq6C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42435.map.gz / Format: CCP4 / Size: 37.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42435.map.gz / Format: CCP4 / Size: 37.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SEN dimer map on the surface of lipid major dimer interface | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.078 Å | ||||||||||||||||||||||||||||||||||||
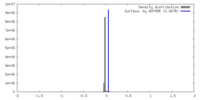
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: SEN dimer map on the surface of lipid combined dimer interfaces
| File | emd_42435_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SEN dimer map on the surface of lipid combined dimer interfaces | ||||||||||||
| Projections & Slices |
| ||||||||||||
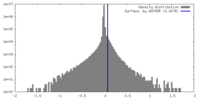
| Density Histograms |
-Half map: SEN dimer half map on the surface of lipid major dimer interface B
| File | emd_42435_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SEN dimer half map on the surface of lipid major dimer interface B | ||||||||||||
| Projections & Slices |
| ||||||||||||
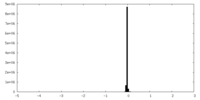
| Density Histograms |
-Half map: SEN dimer half map on the surface of lipid major dimer interface A
| File | emd_42435_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | SEN dimer half map on the surface of lipid major dimer interface A | ||||||||||||
| Projections & Slices |
| ||||||||||||
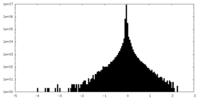
| Density Histograms |
- Sample components
Sample components
-Entire : Streptococcal surface enolase major interface embedded into the s...
| Entire | Name: Streptococcal surface enolase major interface embedded into the surface of a lipid vesicle |
|---|---|
| Components |
|
-Supramolecule #1: Streptococcal surface enolase major interface embedded into the s...
| Supramolecule | Name: Streptococcal surface enolase major interface embedded into the surface of a lipid vesicle type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
| Molecular weight | Theoretical: 95 KDa |
-Macromolecule #1: Enolase
| Macromolecule | Name: Enolase / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: phosphopyruvate hydratase |
|---|---|
| Source (natural) | Organism:  Streptococcus pyogenes (bacteria) Streptococcus pyogenes (bacteria) |
| Molecular weight | Theoretical: 47.543281 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HMSIITDVYA REVLDSRGNP TLEVEVYTES GAFGRGMVPS GASTGEHEAV ELRDGDKSRY LGLGTQKAVD NVNNIIAEAI IGYDVRDQQ AIDRAMIALD GTPNKGKLGA NAILGVSIAV ARAAADYLEV PLYTYLGGFN TKVLPTPMMN IINGGSHSDA P IAFQEFMI ...String: HMSIITDVYA REVLDSRGNP TLEVEVYTES GAFGRGMVPS GASTGEHEAV ELRDGDKSRY LGLGTQKAVD NVNNIIAEAI IGYDVRDQQ AIDRAMIALD GTPNKGKLGA NAILGVSIAV ARAAADYLEV PLYTYLGGFN TKVLPTPMMN IINGGSHSDA P IAFQEFMI MPVGAPTFKE GLRWGAEVFH ALKKILKERG LVTAVGDEGG FAPKFEGTED GVETILKAIE AAGYEAGENG IM IGFDCAS SEFYDKERKV YDYTKFEGEG AAVRTSAEQV DYLEELVNKY PIITIEDGMD ENDWDGWKVL TERLGKRVQL VGD DFFVTN TEYLARGIKE NAANSILIKV NQIGTLTETF EAIEMAKEAG YTAVVSHRSG ETEDSTIADI AVATNAGQIK TGSL SRTDR IAKYNQLLRI EDQLGEVAQY KGIKSFYNLK K UniProtKB: Enolase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Material: GOLD / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 29.0 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 81000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)